Statistical analysis
AGC, AY
23 3 2021
Last updated: 2023-08-29
Checks: 7 0
Knit directory: DEPDC5_D62_Analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220808) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9dd12f1. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Ignored: output/D62_GOresWGCNA.xlsx
Ignored: output/D62_ResTabs_WGCNA.RData
Ignored: output/D62_WGCNA_adj_TOM.RData
Ignored: output/D62_dds_matrix.RData
Ignored: output/D62_mdsplots.RData
Ignored: output/Restab_D62_NTCnoRAPA_DIFF_RAPA.xlsx
Ignored: output/Restab_D62_NTCnoRAPA_DIFF_noRAPA.xlsx
Ignored: output/Restab_D62_NTCnoRAPA_noDIFF_RAPA.xlsx
Ignored: output/Restab_D62_NTCnoRAPA_noDIFF_noRAPA.xlsx
Ignored: output/Restab_D62_NTCwRAPA_DIFF_RAPA.xlsx
Ignored: output/Restab_D62_NTCwRAPA_noDIFF_RAPA.xlsx
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/05_D62_Statistical_analysis.Rmd) and HTML
(docs/05_D62_Statistical_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9dd12f1 | achiocch | 2023-08-29 | wflow_publish(c("./analysis/", "code/"), all = T) |
| html | 9dd12f1 | achiocch | 2023-08-29 | wflow_publish(c("./analysis/", "code/"), all = T) |
| html | 20e7956 | achiocch | 2023-07-31 | Build site. |
| Rmd | 78ac7a6 | achiocch | 2023-07-28 | updated publication figures |
| Rmd | 6cbb9a0 | achiocch | 2023-07-25 | plots fro publication added |
| Rmd | c1f2468 | achiocch | 2022-10-11 | sets the installation procedure in the readme |
| Rmd | 9adf10f | achiocch | 2022-10-11 | solves merge conflict |
| Rmd | dbc34fc | achiocch | 2022-10-11 | minor edits for paper figures |
| Rmd | a7c5f57 | achiocch | 2022-10-10 | fix intaller |
| html | a7c5f57 | achiocch | 2022-10-10 | fix intaller |
| Rmd | 3f72d3a | Andreas Geburtig-Chiocchetti | 2022-08-26 | full analysis pre manuscript version |
| html | 3f72d3a | Andreas Geburtig-Chiocchetti | 2022-08-26 | full analysis pre manuscript version |
| html | e5f1dd3 | achiocch | 2022-08-26 | Build site. |
| Rmd | 4b9fb04 | achiocch | 2022-08-26 | wflow_publish(c("./*")) |
| html | 4b9fb04 | achiocch | 2022-08-26 | wflow_publish(c("./*")) |
| Rmd | 974c39d | achiocch | 2022-08-18 | wflow_publish(c("./*")) |
| html | 974c39d | achiocch | 2022-08-18 | wflow_publish(c("./*")) |
| html | dc78d32 | Andreas Geburtig-Chiocchetti | 2022-08-09 | full analysis pre manuscript version |
| Rmd | f249225 | achiocch | 2022-08-08 | adds data and code |
home = getwd()
knitr::opts_chunk$set(fig.height=6, fig.width=8)Statistical analysis
Which genes are differentially regualted upon KO of DEPDC5
Modelling the KO in all different combinations
if(reanalyze | !file.exists(paste0(home,"/output/D62_ResTabs_KO.RData"))){
## comparisons against noRAPA NTC
Rapamycin=c("noRAPA", "RAPA")
Differentiation=c("noDIFF", "DIFF")
Type_sgRNA<-c("sg2.1","sg2.2")
target="KO"
r = Rapamycin[1]
d = Differentiation[1]
Tp = Type_sgRNA[[1]]
# no random effects included
for(r in Rapamycin){
Rapafilter = SampleInfo$RAPA %in% r
for(d in Differentiation){
Difffilter = SampleInfo$DIFF %in% d
for(Tp in Type_sgRNA){
sgRNAfilter = SampleInfo$gRNA %in% Tp
vs_label=paste0(c("sgNTC", Tp), sep="", collapse="_")
Set = rownames(SampleInfo)[Rapafilter&Difffilter&
sgRNAfilter]
Set = c(Set, rownames(SampleInfo)[SampleInfo$RAPA == "noRAPA" & Difffilter&
SampleInfo$gRNA == "sgNTC"])
lab = paste("restab", "D62_NTCnoRAPA", vs_label, d,r, sep="_")
print(lab)
assign(lab, comparison(ddsMat, Set, target, randomeffect = NULL))
}
}
}
# calculate all combinations against same NTC (treated)
Type_sgRNA<-list(c("sgNTC","sg2.1"),c("sgNTC","sg2.2"))
Cl<-"D62"
# no random effects included
for(r in Rapamycin){
Rapafilter = SampleInfo$RAPA %in% r
for(d in Differentiation){
Difffilter = SampleInfo$DIFF %in% d
for(Tp in Type_sgRNA){
sgRNAfilter = SampleInfo$gRNA %in% Tp
vs_label=paste0(Tp, sep="", collapse="_")
Set = rownames(SampleInfo)[Rapafilter&Difffilter&
sgRNAfilter]
lab = paste("restab", "D62_NTCwRAPA",vs_label, d,r, sep="_")
print(lab)
assign(lab,
comparison(ddsMat, samples = Set,
target =target,randomeffect = c()))
}
}
}
comparisons= apropos("restab")
save(list = comparisons, file = paste0(home,"/output/D62_ResTabs_KO.RData"))
} else
load(file = paste0(home,"/output/D62_ResTabs_KO.RData"))[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA"
[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_noRAPA"
[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA"
[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_noRAPA"
[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_RAPA"
[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_RAPA"
[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_RAPA"
[1] "restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_RAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.1_noDIFF_noRAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.2_noDIFF_noRAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.1_DIFF_noRAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.2_DIFF_noRAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.1_noDIFF_RAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.2_noDIFF_RAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.1_DIFF_RAPA"
[1] "restab_D62_NTCwRAPA_sgNTC_sg2.2_DIFF_RAPA"samesign <- function(x) {abs(sum(sign(x)))==length(x)}
mypval=0.05
colors <- rev(colorRampPalette(brewer.pal(9, "Spectral"))(255))
genelists=lapply(list.files(paste0(home,"/data/genelists_to_test"), full.names = T),
function(x){read.table(x, header=T)})
names(genelists) = gsub(".txt", "", list.files(paste0(home,"/data/genelists_to_test")))
getlinmodoutput = function(targetline="D62",
targetdiff,
targetrapa,
refset="NTCnoRAPA"){
if(refset=="NTCnoRAPA"){
samplesincl=SampleInfo$DIFF==targetdiff &
SampleInfo$RAPA==targetrapa &
SampleInfo$KO == "KO"
samplesincl = samplesincl | (SampleInfo$DIFF==targetdiff &
SampleInfo$RAPA=="noRAPA" &
SampleInfo$KO == "WT")
} else {
samplesincl = SampleInfo$DIFF==targetdiff &
SampleInfo$RAPA==targetrapa
}
pvalrep= get(paste0("restab_",
targetline,"_",refset, "_sgNTC_sg2.1_",
targetdiff, "_" ,
targetrapa))$padj<mypval &
get(paste0("restab_",
targetline, "_",refset,"_sgNTC_sg2.2_",
targetdiff, "_" ,
targetrapa))$padj<mypval
betarep = apply(cbind(get(paste0("restab_",
targetline,"_",refset, "_sgNTC_sg2.1_",
targetdiff, "_" ,
targetrapa))$log2FoldChange,
get(paste0("restab_",
targetline,"_",refset, "_sgNTC_sg2.2_",
targetdiff, "_" ,
targetrapa))$log2FoldChange), 1,
samesign)
idx=which(betarep & pvalrep)
hits=geneids[idx]
restab=data.frame(
entrezgene = geneids[idx],
genname=rowData(ddsMat)[hits,"hgnc"],
log2FC_2.1 = get(paste0("restab_",
targetline,"_",refset, "_sgNTC_sg2.1_",
targetdiff, "_" ,
targetrapa))$log2FoldChange[idx],
fdr_2.1 = get(paste0("restab_",
targetline,"_",refset, "_sgNTC_sg2.1_",
targetdiff, "_" ,
targetrapa))$padj[idx],
log2FC_2.2 = get(paste0("restab_",
targetline,"_",refset, "_sgNTC_sg2.2_",
targetdiff, "_" ,
targetrapa))$log2FoldChange[idx],
fdr_2.2 = get(paste0("restab_",
targetline,"_",refset, "_sgNTC_sg2.2_",
targetdiff, "_" ,
targetrapa))$padj[idx])
print(restab)
write.xlsx(restab, file=paste0(output, "/Restab_",
targetline,"_",refset, "_",
targetdiff, "_",
targetrapa, ".xlsx"))
SamplesSet=SampleInfo[samplesincl,]
plotmatrix = log_2cpm[hits,rownames(SamplesSet)]
rownames(SamplesSet)=SamplesSet$label_rep
colnames(plotmatrix)=SamplesSet$label_rep
rownames(plotmatrix)=rowData(ddsMat)[hits,"hgnc"]
gRNAcol = Dark8[c(1:nlevels(SampleInfo$gRNA))+nlevels(SampleInfo$CellLine)]
names(gRNAcol) = levels(SampleInfo$gRNA)
gRNAcol = gRNAcol[as.character(unique(SamplesSet$gRNA))]
diffcol = brewer.pal(3,"Set1")[1:nlevels(SampleInfo$DIFF)]
names(diffcol) = levels(SampleInfo$DIFF)
diffcol = diffcol[as.character(unique(SamplesSet$DIFF))]
rapacol = brewer.pal(3,"Set2")[1:nlevels(SampleInfo$RAPA)]
names(rapacol) = levels(SampleInfo$RAPA)
rapacol = rapacol[as.character(unique(SamplesSet$RAPA))]
ann_colors = list(
DIFF = diffcol,
RAPA = rapacol,
gRNA = gRNAcol)
collabels = SamplesSet[,c("DIFF","RAPA", "gRNA")] %>%
mutate_all(as.character) %>% as.data.frame()
rownames(collabels)=SamplesSet$label_rep
idx=order(SamplesSet$gRNA)
pheatmap(plotmatrix[,idx],
border_color = NA,
annotation_col = collabels[idx,],
show_rownames = F, show_colnames = F,
annotation_colors = ann_colors,
clustering_method = "ward.D2",
cluster_cols = F,
col = colors,
scale = "row",
main = paste("Normalized log2 counts", targetline, targetdiff, targetrapa))
gene_univers = rownames(ddsMat)
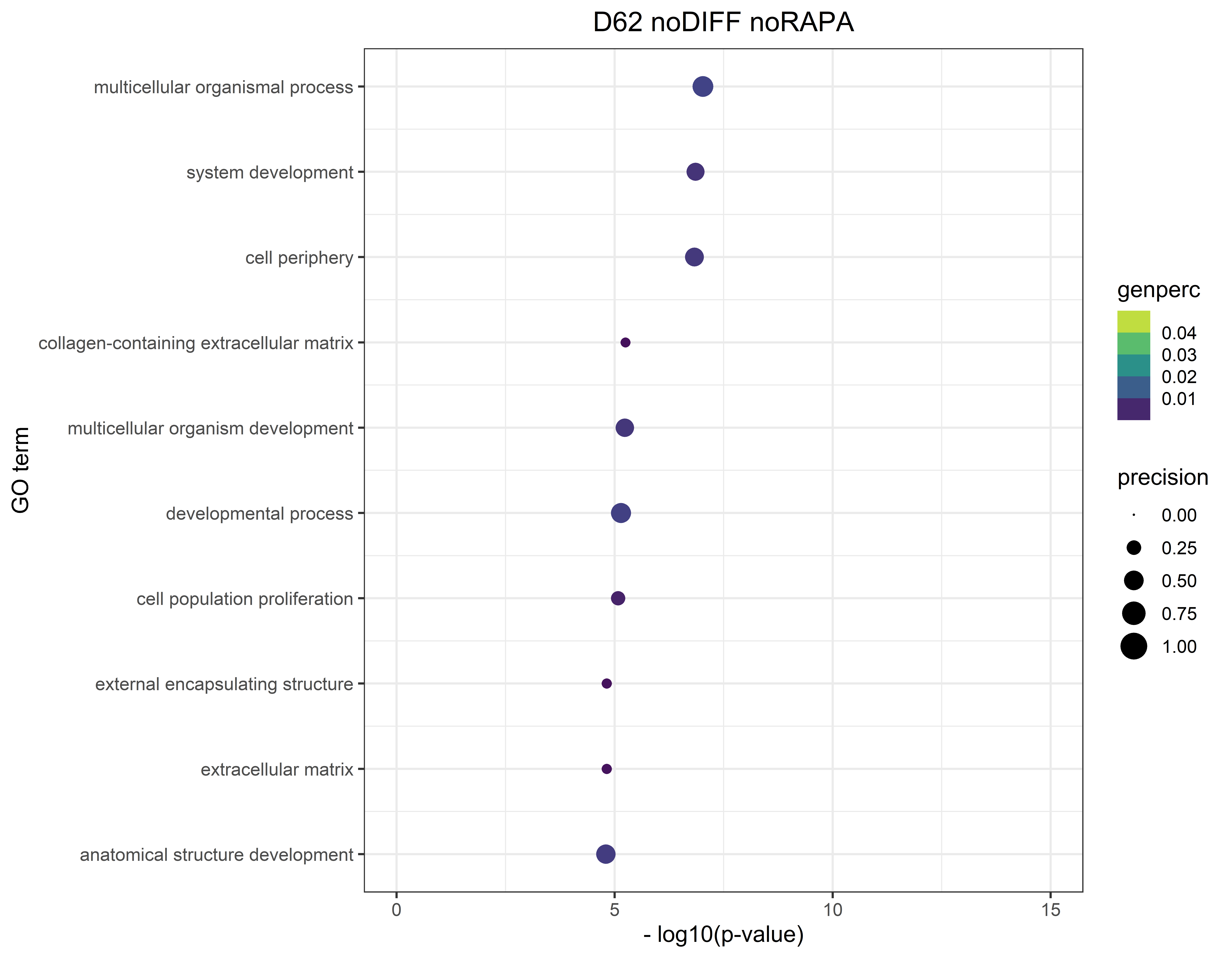
gostres = getGOresults(hits, gene_univers, evcodes = T)
toptab = gostres$result
write.xlsx(toptab, file = paste0(output, "/GOres_",targetline,targetdiff,targetrapa,".xlsx"), sheetName = "GO_enrichment")
idx = grepl("GO|KEGG", toptab$source)
titlename = paste(targetline,targetdiff,targetrapa)
if(!any(idx)){
p = ggplot() + annotate("text", x = 4, y = 25, size=4,
label = "no significant GO term") +
ggtitle(titlename)+theme_void()+
theme(plot.title = element_text(hjust = 0.5))
} else {
p=GOplot(toptab[idx, ], 10, Title = titlename)
}
print(p)
## enrichment test for genelist
Resall=data.frame()
for (gl in names(genelists)){
tmp=table(signif=geneids %in% hits, targetgene=geneids %in% genelists[[gl]]$ENTREZ_ID)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resall = rbind(Resall, res)
}
colnames(Resall)=c("OR", "CI95L", "CI95U", "P")
rownames(Resall)=names(genelists)
Resall$Beta = log(Resall$OR)
Resall$SE = (log(Resall$OR)-log(Resall$CI95L))/1.96
Resall$Padj=p.adjust(Resall$P, method = "bonferroni")
print(Resall)
multiORplot(Resall, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE",
pheno=paste(targetline, targetdiff, targetrapa), xlims=c(-4.5,4.5))
}
geneids=rownames(restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA)KO effect in noDIFF noRAPA
D62
getlinmodoutput(targetdiff = "noDIFF",targetrapa = "noRAPA") entrezgene genname log2FC_2.1 fdr_2.1 log2FC_2.2 fdr_2.2
1 64856 VWA1 1.0892844 3.075562e-12 0.7285097 3.335990e-06
2 26038 CHD5 2.6315908 1.417316e-07 1.4609239 1.668736e-02
3 3925 STMN1 -0.8131038 6.841853e-11 -0.4047316 1.617550e-04
4 90853 SPOCD1 1.5410330 1.306139e-05 0.9545620 4.769198e-02
5 23499 MACF1 0.3837815 4.965528e-02 0.3341228 2.673200e-02
6 59269 HIVEP3 0.8406193 5.691220e-03 0.7731647 8.991416e-04
7 11004 KIF2C -1.4716331 3.774881e-06 -1.2753566 6.553854e-08
8 5052 PRDX1 -0.5761057 1.440015e-09 -0.4282492 7.515508e-09
9 9077 DIRAS3 1.8022491 1.054607e-14 1.5352770 9.581298e-21
10 81849 ST6GALNAC5 -0.7856164 1.709336e-10 -0.4114943 1.159554e-05
11 64123 ADGRL4 1.3768823 1.525051e-03 1.8273933 5.105453e-10
12 2633 GBP1 4.3457775 1.595376e-04 4.0102130 9.650536e-05
13 84722 PSRC1 -0.4534422 9.730923e-04 -0.3020121 9.492046e-03
14 3321 IGSF3 -0.8433839 9.585479e-04 -0.5473542 6.989319e-03
15 10628 TXNIP -1.3810106 4.071429e-05 -1.1473212 4.686387e-07
16 140576 S100A16 0.8919626 1.093717e-09 0.3317805 4.172758e-02
17 10763 NES 0.7606565 1.467078e-13 0.4262272 8.725999e-07
18 57863 CADM3 -2.0260731 1.510930e-03 -1.0777568 3.583125e-02
19 400793 C1orf226 0.8127671 6.400278e-03 0.6002242 2.740678e-02
20 4921 DDR2 -0.8525309 3.949671e-02 -0.8846002 8.610829e-04
21 5999 RGS4 2.1857048 4.032092e-08 2.0257520 1.248142e-07
22 6004 RGS16 1.3232831 2.664105e-11 0.5609847 2.313075e-02
23 64710 NUCKS1 -0.4835910 2.329196e-06 -0.2913098 1.451638e-03
24 1063 CENPF -1.1466949 3.266629e-13 -0.4548584 2.238609e-04
25 149111 CNIH3 1.0690978 4.325986e-03 1.0625651 4.806234e-04
26 84886 C1orf198 0.5484394 3.527610e-02 0.5158203 2.465076e-02
27 4811 NID1 -1.5129348 2.124236e-03 -1.0116004 2.309960e-02
28 22944 KIN -2.0055566 5.757344e-04 -1.1838012 1.061975e-02
29 5660 PSAP 0.7111062 4.410466e-20 0.1798656 8.451417e-03
30 54541 DDIT4 1.0557531 3.490589e-03 0.8450571 2.198764e-02
31 5328 PLAU 1.2384477 1.577948e-02 1.0758169 3.641623e-02
32 84858 ZNF503 1.8245316 4.091039e-04 1.5504984 1.376196e-03
33 3988 LIPA 1.0234270 1.371967e-04 0.6509579 3.729125e-02
34 143282 FGFBP3 0.6243988 2.721432e-04 0.4629851 4.527298e-03
35 26509 MYOF 1.7145925 3.329288e-02 2.0874494 1.456813e-03
36 9124 PDLIM1 1.1001337 7.354962e-04 1.1004307 2.870465e-06
37 83742 MARVELD1 -0.8431712 1.326637e-02 -0.6517105 3.315254e-02
38 6319 SCD 0.5331130 1.645543e-05 0.3371696 1.108394e-03
39 6468 FBXW4 1.1779903 1.481301e-03 0.7718936 3.641623e-02
40 259217 HSPA12A 1.8421463 3.370295e-03 1.6193468 2.292344e-02
41 642938 INSYN2A 0.4027642 4.015934e-02 0.7777285 6.605916e-10
42 4288 MKI67 -1.0123111 1.442152e-06 -0.5893857 5.819468e-03
43 977 CD151 1.2635289 3.728767e-27 0.3295937 9.815768e-03
44 6240 RRM1 -0.8816314 1.153480e-02 -1.0401431 1.908546e-04
45 4004 LMO1 -2.3567118 2.079132e-07 -2.7131580 1.254653e-21
46 10553 HTATIP2 1.9333095 1.690227e-02 2.0019304 5.655760e-04
47 5080 PAX6 -0.9181084 1.019508e-02 -0.8147835 2.564557e-03
48 960 CD44 1.4591708 9.338179e-16 1.1683319 1.210130e-15
49 79026 AHNAK 1.5851868 1.544050e-09 0.9169080 4.768239e-04
50 1717 DHCR7 0.7307510 3.188177e-04 0.3960401 3.001859e-02
51 26011 TENM4 1.7246307 2.481137e-02 1.5154269 4.675757e-02
52 11098 PRSS23 0.8798684 1.960728e-02 0.9004492 3.761899e-04
53 1410 CRYAB 1.1173770 3.476939e-12 0.5931808 1.088950e-04
54 6876 TAGLN 0.6543144 1.983800e-13 0.3420865 6.018107e-08
55 1656 DDX6 -0.4617054 2.166969e-02 -0.3694408 5.537409e-03
56 2012 EMP1 0.6479519 1.563592e-02 1.1887114 1.119352e-12
57 196410 METTL7B 1.5599602 3.020553e-12 0.7182732 2.255546e-03
58 967 CD63 0.9019072 3.851592e-39 0.2540967 1.196946e-05
59 4035 LRP1 0.6449247 4.076243e-05 0.5918438 1.866013e-07
60 4193 MDM2 0.6719346 3.844160e-05 0.3615605 1.262560e-02
61 160335 TMTC2 -1.6135376 6.014427e-06 -1.0544925 1.724845e-04
62 490 ATP2B1 -0.7878372 1.474724e-03 -0.6061956 4.808735e-04
63 5829 PXN 0.7640621 4.062369e-04 0.5820746 7.936104e-03
64 3146 HMGB1 -0.7285274 5.966944e-16 -0.3215517 6.194211e-07
65 8848 TSC22D1 0.5613517 5.123576e-04 0.6635049 1.163073e-11
66 8660 IRS2 0.4101733 2.430542e-05 0.2984944 4.872851e-04
67 28526 <NA> 1.9278887 1.741341e-07 1.9178656 1.968941e-11
68 54916 TMEM260 -1.0306993 3.988816e-02 -0.9104398 3.976121e-02
69 57570 TRMT5 -0.6824356 9.703832e-05 -0.3831739 6.823688e-03
70 23224 SYNE2 -1.3811382 1.909678e-06 -0.7362985 6.120540e-05
71 64093 SMOC1 0.7201019 1.536550e-07 0.3597993 9.699354e-04
72 9369 NRXN3 -1.7174772 2.192983e-05 -1.0877059 8.093515e-04
73 55384 MEG3 1.6246309 6.353745e-22 1.1918119 3.931182e-15
74 113146 AHNAK2 2.2298981 1.100829e-04 2.6577306 1.642724e-14
75 11245 GPR176 2.1616993 3.793467e-03 2.0977492 1.717827e-03
76 90417 KNSTRN -1.3213590 3.768730e-03 -0.8170137 4.172758e-02
77 51203 NUSAP1 -1.3682393 1.819314e-06 -0.5193085 8.702281e-03
78 2628 GATM 0.6525196 3.474952e-02 1.0416366 7.594344e-12
79 302 ANXA2 0.3964197 1.975148e-04 0.2995517 1.056447e-02
80 7168 TPM1 0.5163549 2.189321e-02 0.4713141 1.858398e-03
81 9768 PCLAF -1.6192503 6.619244e-04 -1.0918606 3.178054e-03
82 3671 ISLR 1.4582038 7.636745e-08 0.8562869 9.938267e-04
83 23205 ACSBG1 0.6910217 1.265573e-02 0.5922977 3.041423e-02
84 9055 PRC1 -0.7454089 1.738144e-02 -0.5070052 4.882398e-02
85 56963 RGMA 1.6179562 2.570150e-22 0.7273235 5.283814e-07
86 23336 SYNM 0.7053394 8.114026e-05 0.7285223 3.606307e-08
87 51330 TNFRSF12A 1.4059052 4.373945e-20 0.9554401 6.348958e-16
88 2013 EMP2 -0.7546354 7.607574e-05 -0.4966014 7.060439e-03
89 26471 NUPR1 2.3612545 1.870745e-12 1.0657975 2.418253e-02
90 4313 MMP2 0.8703997 3.440811e-09 0.4221113 6.487195e-03
91 6376 CX3CL1 0.8954558 1.512042e-03 0.5519566 1.955410e-02
92 794 CALB2 -4.6167606 1.822472e-06 -2.1172122 2.112181e-03
93 23406 COTL1 0.7992038 1.400921e-09 0.4700650 1.672984e-06
94 79007 DBNDD1 1.9205874 2.565613e-24 0.6772243 1.108394e-03
95 9212 AURKB -1.2399686 2.287452e-03 -1.1963693 1.632320e-03
96 9423 NTN1 1.4854613 2.618750e-12 0.6558489 6.669248e-03
97 9953 HS3ST3B1 -0.7862300 1.911337e-04 -0.4187471 7.670737e-03
98 5376 PMP22 0.5572320 3.108196e-03 0.4802813 1.670511e-03
99 125144 SNHG29 -0.5331262 6.565905e-03 -0.2835938 2.475929e-02
100 7153 TOP2A -0.9721903 1.993781e-11 -0.8239175 1.222927e-15
101 284119 CAVIN1 1.0611724 1.125087e-04 0.6185919 4.656529e-02
102 2670 GFAP 1.0690762 1.553293e-30 0.2604783 4.641576e-03
103 6426 SRSF1 -0.9521000 2.684500e-06 -0.5783965 6.608999e-04
104 54549 SDK2 -0.9445922 4.684966e-02 -0.8412877 4.125061e-02
105 283991 UBALD2 0.5569752 3.228727e-05 0.3796500 4.175454e-03
106 332 BIRC5 -1.2781479 4.572826e-04 -0.7416046 8.606921e-04
107 284184 NDUFAF8 0.8494269 4.400209e-12 0.3473532 8.122019e-03
108 11031 RAB31 0.3245603 2.321577e-03 0.2509527 1.338585e-02
109 1000 CDH2 0.2478272 2.666336e-02 0.3194867 5.502703e-08
110 115106 HAUS1 -1.6973970 1.660721e-03 -0.9655808 8.649361e-03
111 4645 MYO5B -1.4814373 9.958677e-03 -0.8092195 3.583125e-02
112 5596 MAPK4 -1.1639881 3.998119e-03 -0.9126168 1.983599e-02
113 976 ADGRE5 1.5563103 2.052428e-10 0.9699747 2.475935e-04
114 9518 GDF15 1.7737014 2.565613e-24 0.9110701 2.968916e-08
115 348 APOE 2.0368060 6.914416e-20 1.2379642 7.171243e-11
116 83987 CCDC8 -2.7519728 5.038789e-04 -2.1441453 2.994825e-04
117 27113 BBC3 1.5017524 3.074687e-09 1.1134730 4.976536e-06
118 23645 PPP1R15A 0.7988109 3.681655e-04 0.5178092 3.288663e-02
119 79784 MYH14 -3.2161023 5.022167e-07 -3.3862033 9.359765e-10
120 151354 LRATD1 0.9973829 3.281705e-09 0.8068247 5.502703e-08
121 6432 SRSF7 -0.5308112 1.770509e-02 -0.4885241 3.027381e-02
122 91461 PKDCC -0.5131360 2.364997e-04 -0.4134268 6.372799e-05
123 25927 CNRIP1 -6.6989984 1.205616e-03 -5.2262001 2.255546e-03
124 3099 HK2 -1.7924230 3.763123e-03 -1.1143942 3.847751e-03
125 343990 CRACDL 1.5513833 9.048284e-04 1.5050862 5.244535e-06
126 164832 LONRF2 0.8062659 1.016761e-03 0.6143445 1.061975e-02
127 4175 MCM6 -1.1984461 1.248926e-02 -1.0400180 6.655388e-03
128 130574 LYPD6 -1.2606232 4.596596e-02 -1.6174273 9.289330e-03
129 114793 FMNL2 0.8390155 1.687899e-08 0.5687998 1.361514e-05
130 55854 ZC3H15 -0.6060614 1.144384e-02 -0.4326721 3.653289e-02
131 26010 SPATS2L 0.4043183 4.131211e-02 0.5234441 7.203596e-04
132 389073 C2orf80 -0.6857721 3.141838e-05 -0.3998037 4.014575e-03
133 1373 CPS1 -2.4069129 3.329915e-02 -1.5377128 1.121948e-02
134 2637 GBX2 0.9261524 4.325986e-03 0.6662947 2.507785e-02
135 10267 RAMP1 1.3725884 8.222159e-12 0.8504061 2.690388e-05
136 5834 PYGB 1.0717781 1.273346e-07 0.6080728 2.456034e-02
137 22974 TPX2 -1.3244518 9.287860e-04 -0.7959339 5.286385e-03
138 1299 COL9A3 0.5763424 4.894524e-05 0.2836497 7.718569e-03
139 3785 KCNQ2 -0.6122229 2.017555e-03 -0.6179499 3.191695e-05
140 140578 CHODL -2.8027100 5.361518e-04 -1.5336391 2.067193e-02
141 116448 OLIG1 0.9341630 1.781013e-05 0.5876127 1.493659e-02
142 5121 PCP4 -1.5677901 7.982074e-03 -1.1880812 4.481359e-02
143 1826 DSCAM 1.5771813 7.354962e-04 2.1020644 2.810046e-11
144 80781 COL18A1 1.3186038 2.371954e-07 0.8089897 6.655388e-03
145 1312 COMT 0.5106568 1.009903e-05 0.3231095 8.796510e-03
146 23467 NPTXR 1.1217928 9.404146e-13 0.5426083 9.571565e-04
147 10752 CHL1 -1.0507620 2.039054e-12 -0.4938486 1.008946e-05
148 23171 GPD1L -1.6672836 8.686360e-04 -1.2531363 8.991416e-04
149 56992 KIF15 -1.1460509 1.350669e-02 -1.3062017 1.097283e-02
150 151887 CCDC80 0.7869097 4.245320e-08 0.5435400 4.786979e-06
151 2596 GAP43 1.6364412 2.243038e-59 1.0798978 1.176119e-32
152 11343 MGLL 0.9208202 4.827230e-05 0.6124697 8.907194e-03
153 5947 RBP1 -0.4676065 2.065758e-02 -0.7465618 2.229788e-07
154 440982 <NA> 3.0724570 3.299840e-02 3.5022415 1.313114e-03
155 10051 SMC4 -1.3310413 1.626927e-13 -0.5801032 1.293223e-03
156 8646 CHRD 1.6869291 3.299840e-02 1.7578236 2.135439e-03
157 6480 ST6GAL1 -4.2851264 1.646800e-02 -7.1414798 4.717064e-04
158 131583 FAM43A 1.7599988 8.970095e-05 1.2640133 5.295856e-03
159 347 APOD -1.9862063 1.760697e-03 -1.9388294 6.526021e-03
160 10460 TACC3 -0.9053632 1.593226e-02 -0.9375377 6.553253e-05
161 80333 KCNIP4 -2.1231152 7.429885e-05 -1.2853303 3.367812e-02
162 5099 PCDH7 -2.6026035 6.185863e-03 -1.5828258 2.013908e-02
163 5156 PDGFRA -0.9551616 3.879820e-08 -0.4385603 4.971439e-04
164 3490 IGFBP7 1.4858305 1.043598e-18 1.8278345 1.747889e-53
165 79966 SCD5 0.4050749 2.026390e-04 0.7008740 2.372241e-28
166 8404 SPARCL1 2.1765723 5.749106e-10 2.1513872 3.918759e-16
167 3015 H2AZ1 -0.7064878 1.621255e-07 -0.4993665 1.898985e-05
168 1062 CENPE -2.1755855 1.275813e-19 -0.5724353 4.658449e-03
169 55013 MCUB -0.8289339 5.110689e-03 -0.6419360 1.166422e-03
170 2983 GUCY1B1 -1.9332928 1.077333e-02 -1.3467286 1.873558e-02
171 4889 NPY5R -1.2088762 3.804174e-02 -1.3279815 1.095295e-03
172 3148 HMGB2 -1.4513006 1.140854e-07 -0.5872440 1.763412e-04
173 55714 TENM3 -2.5344066 2.026891e-13 -0.5553902 2.432824e-02
174 1767 DNAH5 5.8165242 3.636635e-02 5.6533364 4.525126e-02
175 7204 TRIO 0.7066729 8.199761e-03 0.5629365 7.524829e-03
176 6507 SLC1A3 -0.5128031 3.890674e-07 -0.3899367 5.506826e-07
177 102467080 <NA> -2.2094626 4.470417e-02 -2.7773677 4.412879e-03
178 728723 <NA> 7.3332529 9.430977e-04 6.3853331 2.150554e-02
179 22836 RHOBTB3 -0.4283172 3.417498e-02 -0.3694922 6.071604e-04
180 4001 LMNB1 -1.2366912 5.892292e-11 -0.6987209 4.365961e-06
181 2201 FBN2 -1.3303425 6.131690e-03 -1.1780917 9.492046e-03
182 10112 KIF20A -1.3658440 8.555787e-05 -1.0750296 4.376420e-04
183 84418 CYSTM1 0.7226909 1.854422e-03 0.4647585 2.967546e-02
184 113829 SLC35A4 1.2350898 5.396466e-06 0.6322482 3.538265e-02
185 57717 PCDHB16 -1.3028842 5.530593e-04 -0.7867778 1.402857e-02
186 2246 FGF1 0.7332113 8.555787e-05 0.6731462 6.551751e-05
187 202374 STK32A 1.6462918 2.485989e-03 1.2999571 6.823688e-03
188 6678 SPARC 0.7430602 6.700525e-26 0.6107188 8.261940e-23
189 9232 PTTG1 -0.6679420 1.723066e-03 -0.5335900 4.553954e-06
190 221749 PXDC1 0.5202452 1.483568e-02 0.4267824 3.943582e-02
191 51299 NRN1 2.8346049 3.048430e-04 2.7340323 1.441315e-04
192 10590 SCGN -3.2959812 5.409653e-03 -1.0164333 2.302360e-06
193 222698 NKAPL 3.5941338 1.499320e-02 3.5831756 2.563656e-04
194 7726 TRIM26 6.5704536 2.168534e-03 6.5387254 2.785842e-03
195 221491 SMIM29 0.9425016 1.718713e-04 0.4821175 4.738456e-02
196 1026 CDKN1A 1.7453810 8.686504e-79 0.8928271 5.940822e-32
197 3910 LAMA4 0.4077226 4.634758e-02 0.4381660 1.336896e-02
198 57224 NHSL1 1.4592479 5.493698e-03 1.2016807 2.723916e-02
199 6196 RPS6KA2 0.8475993 3.714411e-07 0.4745065 4.658449e-03
200 221833 SP8 -0.9898680 5.396466e-06 -0.6132468 1.683932e-03
201 55536 CDCA7L -0.7780624 2.352479e-04 -0.5031204 1.644441e-02
202 10457 GPNMB 1.5160771 1.748388e-04 1.2495676 9.793869e-04
203 4852 NPY -1.6635528 2.160922e-08 -1.3049255 1.069403e-07
204 64409 GALNT17 2.4304410 4.418133e-06 1.4306199 3.153610e-02
205 23089 PEG10 0.3634026 1.838462e-02 0.5073602 9.958056e-09
206 5054 SERPINE1 1.2476521 9.029955e-15 1.2030681 1.863591e-18
207 5764 PTN 0.3462228 1.652673e-03 0.3256584 8.725999e-07
208 8795 TNFRSF10B 0.7801657 2.767747e-02 0.8052392 2.038249e-03
209 4741 NEFM 1.8245160 2.718716e-04 1.4768708 1.675617e-03
210 4747 NEFL 1.3043536 1.356134e-07 0.8373609 5.766403e-04
211 2185 PTK2B 1.8432074 1.983800e-13 1.0401362 8.117528e-05
212 1191 CLU 0.6346144 2.842432e-18 0.3188940 5.764001e-10
213 157570 ESCO2 -2.3392823 2.150740e-02 -1.5660868 9.836713e-04
214 5327 PLAT 0.5763119 4.478844e-04 0.4540847 1.497045e-03
215 10434 LYPLA1 -1.2302889 1.589568e-03 -0.6142569 3.976121e-02
216 11075 STMN2 -4.7688923 1.772183e-02 -4.3525251 1.838981e-02
217 5375 PMP2 -1.3010953 1.823070e-46 -0.2563026 2.613947e-03
218 1345 COX6C 0.4028032 2.288205e-04 0.2365620 2.616200e-03
219 29028 ATAD2 -1.3185527 6.489143e-04 -0.8523916 1.121948e-02
220 114907 FBXO32 0.7502901 1.563154e-02 0.9131897 8.216921e-06
221 169044 COL22A1 1.0208083 8.337346e-03 0.9880348 1.443547e-03
222 340371 NRBP2 0.8884357 4.930026e-06 0.5513259 4.513721e-02
223 11168 PSIP1 -0.7564217 5.843104e-05 -0.3478707 1.767618e-02
224 6194 RPS6 -0.4727294 1.536550e-07 -0.1956618 2.833713e-02
225 9568 GABBR2 1.8836545 4.228593e-19 1.7690687 2.524294e-26
226 10592 SMC2 -1.5498404 8.635201e-07 -0.7496846 1.262560e-02
227 2934 GSN 0.9595896 3.928363e-03 0.8539323 7.811367e-05
228 64855 NIBAN2 1.0039742 4.038707e-07 0.4461762 1.575051e-02
229 56262 LRRC8A 1.0113393 4.043294e-15 0.3689812 6.436992e-03
230 5730 PTGDS 1.3860170 1.983800e-13 0.7788765 2.690388e-05
231 4571 <NA> 0.9828721 7.409716e-07 0.8379026 2.855739e-06
232 100189191 <NA> -6.6852030 1.335824e-02 -5.7268208 3.719196e-02
233 51186 TCEAL9 0.9506406 8.102250e-13 0.3986340 5.340717e-05
234 27018 BEX3 0.2314104 1.607417e-02 0.2416858 1.629642e-04
235 1641 DCX -0.8989459 1.739980e-07 -0.7034328 1.196946e-05
236 5358 PLS3 0.6070451 1.377846e-02 0.7926681 6.018107e-08
237 10178 TENM1 -1.2341158 6.170708e-04 -1.1153249 2.026317e-05Lade nötiges Paket: gprofiler2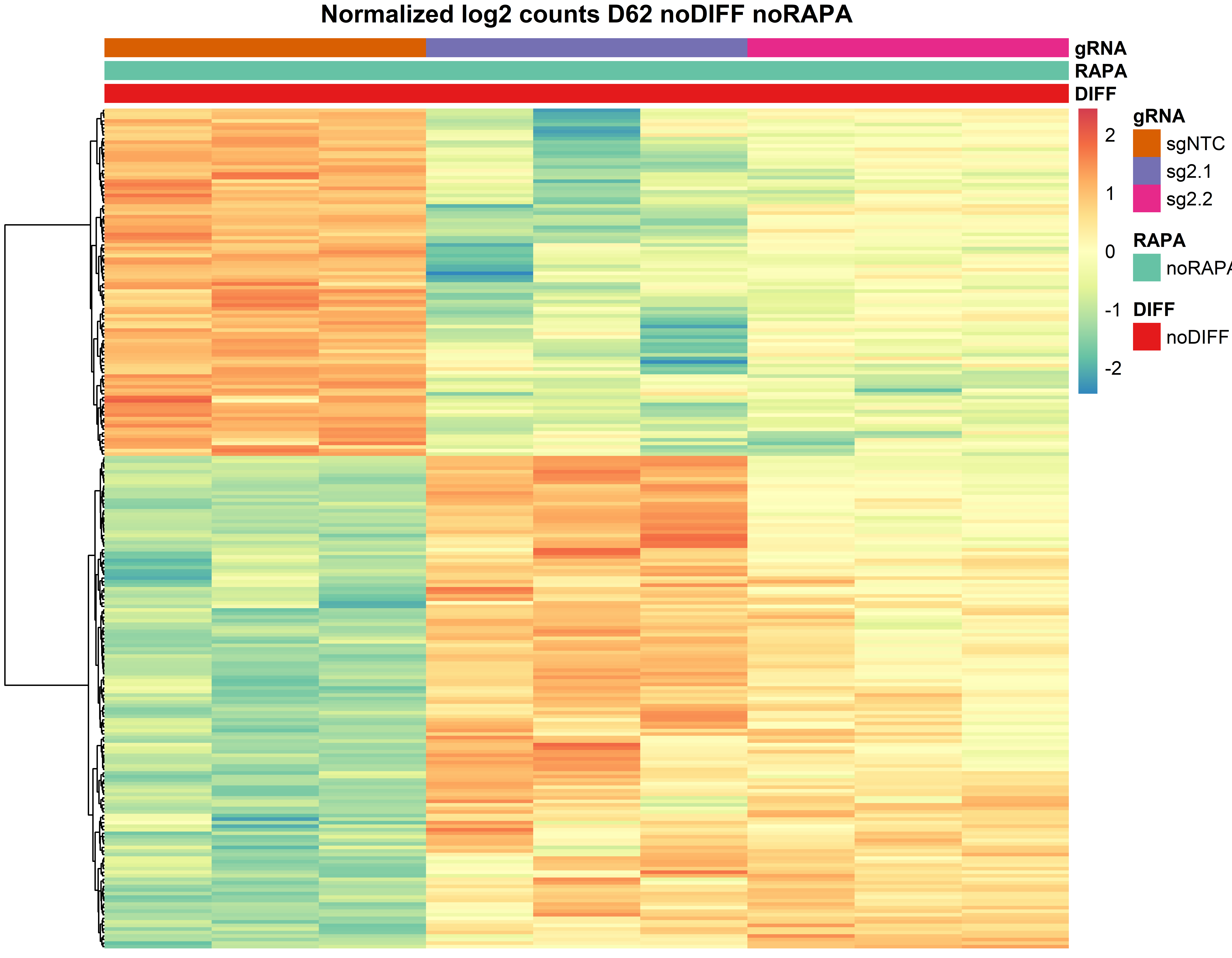
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.2493170 0.71100298 2.060004 3.997515e-01
02_TSC2_KO_Grabole_2016 1.7532671 1.34304311 2.286689 2.358088e-05
03_TSC2_Grabole-2016 1.9083166 1.44871521 2.528768 2.001931e-06
04_TSC2-tuber-Martin2017 3.9177364 1.52014793 8.493193 3.102625e-03
05_TSC2-SEN_SEGA-Martin2017 3.3787094 2.54592258 4.458609 3.086603e-16
06_all_EpilepsyGene 0.9136922 0.32971921 2.038430 1.000000e+00
07_high_conf_EpilepsyGene 0.9328263 0.11105192 3.480452 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene 2.0509392 0.54469645 5.469632 1.409101e-01
09_Epi25_dataset 0.6846692 0.01705876 3.958973 1.000000e+00
10_SFARI_all 1.4590930 0.52462921 3.272844 3.166033e-01
11_SFARI_score4plus 1.2990334 0.34713723 3.430442 5.563158e-01
12_DeRubeis_RDNV 0.7195488 0.01791718 4.166238 1.000000e+00
13_Iossifov_RDNV 1.8931988 1.05502770 3.174616 2.247509e-02
14_FMRP_Darnell 1.4892943 0.87449385 2.399380 1.134977e-01
15_Voineagu_M12 0.4945257 0.10074334 1.475620 2.930041e-01
16_Voineagu_M16 2.9424386 1.63314424 4.959172 2.803166e-04
17_ID_genes_Neel 0.8929799 0.28525539 2.136302 1.000000e+00
18_ID_genes_pinto2014 0.4781289 0.05723250 1.765901 4.470429e-01
19_Fromer_SCZ_FunDN 1.4522062 0.73400740 2.611490 2.128794e-01
20_Cocchi_Pruning 3.3992671 1.06620904 8.361197 1.971963e-02
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.22259701 0.2875896 1.000000e+00
02_TSC2_KO_Grabole_2016 0.56148098 0.1359913 4.716177e-04
03_TSC2_Grabole-2016 0.64622152 0.1405839 4.003861e-05
04_TSC2-tuber-Martin2017 1.36551403 0.4830135 6.205249e-02
05_TSC2-SEN_SEGA-Martin2017 1.21749380 0.1443881 6.173205e-15
06_all_EpilepsyGene -0.09026153 0.5200267 1.000000e+00
07_high_conf_EpilepsyGene -0.06953629 1.0858271 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene 0.71829783 0.6764410 1.000000e+00
09_Epi25_dataset -0.37881944 1.8838123 1.000000e+00
10_SFARI_all 0.37781502 0.5218768 1.000000e+00
11_SFARI_score4plus 0.26162044 0.6732936 1.000000e+00
12_DeRubeis_RDNV -0.32913090 1.8841145 1.000000e+00
13_Iossifov_RDNV 0.63826786 0.2983168 4.495017e-01
14_FMRP_Darnell 0.39830237 0.2716390 1.000000e+00
15_Voineagu_M12 -0.70415611 0.8117465 1.000000e+00
16_Voineagu_M16 1.07923870 0.3003732 5.606332e-03
17_ID_genes_Neel -0.11319125 0.5822342 1.000000e+00
18_ID_genes_pinto2014 -0.73787502 1.0830400 1.000000e+00
19_Fromer_SCZ_FunDN 0.37308394 0.3481225 1.000000e+00
20_Cocchi_Pruning 1.22355984 0.5915563 3.943925e-01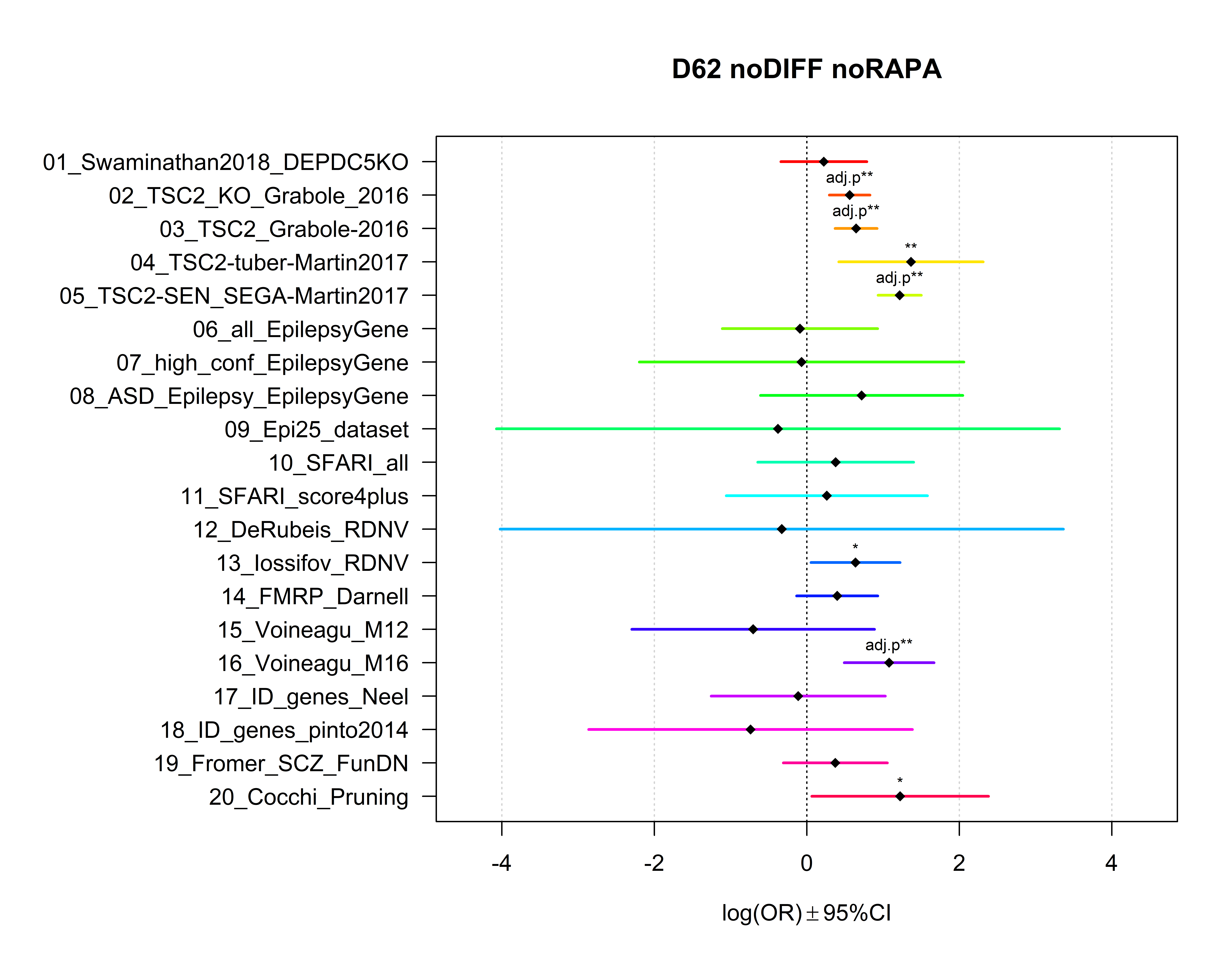
KO effect in DIFF noRAPA
D62
getlinmodoutput(targetdiff = "DIFF",targetrapa = "noRAPA") entrezgene genname log2FC_2.1 fdr_2.1 log2FC_2.2 fdr_2.2
1 375790 AGRN -0.4095366 4.541936e-02 -0.8193766 2.940741e-03
2 51150 SDF4 -0.5045009 2.016520e-03 -0.6130317 1.126153e-04
3 64856 VWA1 -0.5711883 2.969308e-03 -1.4292346 5.573565e-15
4 1953 MEGF6 -1.0513995 1.667149e-02 -2.6636915 7.100257e-05
5 26038 CHD5 -1.2188855 8.890761e-03 -1.2908928 2.557452e-02
6 5351 PLOD1 -0.6033686 2.041058e-03 -0.5114490 3.982798e-02
7 9249 DHRS3 -1.3190383 8.751526e-16 -1.2467008 2.728191e-18
8 126917 IFFO2 -0.3809991 4.730754e-02 -1.2535537 8.070086e-09
9 2048 EPHB2 -0.8733756 2.142055e-09 -1.1159219 8.225789e-13
10 3925 STMN1 0.7854586 3.992855e-07 1.4092446 4.302212e-25
11 2537 IFI6 -0.8526948 2.873716e-06 -0.8392726 1.082687e-04
12 10657 KHDRBS1 0.4607051 7.854427e-03 0.4414765 3.164186e-02
13 65108 MARCKSL1 0.4911297 4.223750e-02 0.9807717 2.059921e-06
14 6421 SFPQ 0.4424978 1.963220e-02 0.4953071 2.274753e-02
15 63967 CLSPN 1.5050166 2.994132e-02 1.6142082 4.662595e-02
16 84879 MFSD2A 0.9674876 1.115952e-05 1.8064180 2.239033e-20
17 6513 SLC2A1 -0.5393673 2.496997e-02 -0.7859054 2.126528e-03
18 1996 ELAVL4 3.2526117 5.784672e-06 4.0471939 5.265819e-09
19 5865 RAB3B 3.5792799 2.097445e-05 4.3609050 4.842433e-08
20 3725 JUN 0.7207107 3.071839e-08 0.5364941 6.557799e-03
21 4774 NFIA 0.5084119 9.704186e-04 0.7115189 2.442476e-05
22 91624 NEXN 0.9933624 1.064809e-03 1.2008241 4.905177e-05
23 3491 CCN1 1.1195767 8.496264e-14 1.2106373 1.267284e-12
24 9411 ARHGAP29 1.7545351 1.866874e-02 1.8531499 3.499818e-02
25 84722 PSRC1 -0.3880493 2.124082e-02 -0.5416116 5.754725e-03
26 3752 KCND3 4.4614872 1.091255e-02 4.7313571 1.538811e-02
27 7482 WNT2B -1.4641738 3.292720e-03 -3.1533870 3.473845e-02
28 54879 ST7L -1.1975888 2.855280e-03 -2.0893903 8.782753e-08
29 389 RHOC -0.7131818 4.100469e-04 -0.8086711 1.375990e-04
30 476 ATP1A1 -1.1090774 4.413887e-09 -0.6351347 7.433223e-03
31 10628 TXNIP -1.0242225 7.601973e-03 -1.5728240 8.349711e-04
32 25832 NBPF14 -0.5964256 1.125473e-02 -0.7179448 7.102298e-03
33 4170 MCL1 0.5791882 9.711034e-03 0.5315779 4.022081e-02
34 10962 MLLT11 0.6716757 4.183809e-03 0.9674580 4.717386e-04
35 6282 S100A11 -0.6710110 1.980465e-04 -1.4509000 1.147719e-08
36 112770 GLMP -1.1638935 1.575170e-04 -0.8287964 1.362657e-02
37 10485 MIR9-1HG 0.3831640 2.381681e-03 0.3859450 2.673569e-02
38 93183 PIGM -0.8618055 1.763717e-03 -0.9515013 1.694960e-03
39 477 ATP1A2 1.4300732 5.252651e-05 2.0030399 1.842601e-09
40 5999 RGS4 1.4755372 1.310525e-02 1.7994122 5.067488e-03
41 387597 ILDR2 -0.7566188 3.030088e-04 -0.9090916 1.155738e-04
42 481 ATP1B1 1.0534462 6.401206e-03 1.2793952 1.469068e-03
43 23057 NMNAT2 3.4078962 9.278183e-04 3.5039038 4.067671e-03
44 23127 COLGALT2 -0.6544929 3.489726e-03 -1.0207651 1.734016e-06
45 5997 RGS2 2.3462296 3.117778e-03 2.3989907 1.226128e-02
46 23046 KIF21B 1.3941358 4.201668e-02 2.8823435 3.849660e-10
47 23612 PHLDA3 -1.0070559 6.558898e-08 -0.5809101 1.902293e-02
48 89796 NAV1 0.9096642 9.397460e-05 1.2045189 2.779659e-08
49 55220 KLHDC8A 0.4221899 4.447089e-02 1.1680073 2.401560e-12
50 254428 SLC41A1 -0.6733399 1.170382e-02 -1.0331474 4.445111e-04
51 50486 G0S2 -1.2865007 3.551702e-02 -3.7678927 6.554199e-37
52 1063 CENPF 0.9584688 1.524609e-04 0.6809078 2.438204e-02
53 3930 LBR 1.1595800 2.660623e-04 1.1648918 1.816828e-03
54 7044 LEFTY2 -1.9062784 1.973434e-02 -5.5053780 1.299284e-06
55 56776 FMN2 1.8126370 2.130849e-05 1.4381718 1.343422e-02
56 5214 PFKP -0.7177737 3.635803e-03 -0.9869597 9.825263e-04
57 1316 KLF6 0.4499054 4.566323e-02 0.9463501 3.670025e-06
58 5209 PFKFB3 0.8932132 1.722147e-06 0.6416209 7.841295e-03
59 56243 KIAA1217 -1.0182112 1.357757e-02 -1.6549404 3.325038e-04
60 57512 GPR158 6.2437538 1.069829e-02 6.1780211 4.217540e-02
61 3185 HNRNPF 0.4833284 1.223805e-02 0.5246720 4.644121e-02
62 118738 ZNF488 -1.3684277 1.512700e-05 -1.5220924 2.643263e-05
63 100507008 <NA> -1.0774683 1.550885e-02 -1.1999015 2.280369e-02
64 84159 ARID5B 1.0976502 1.040875e-09 0.7005891 1.321530e-03
65 55680 RUFY2 1.4578324 2.486895e-02 1.7756965 1.200570e-02
66 140766 ADAMTS14 -1.0682409 2.477355e-02 -1.9481264 1.072294e-04
67 64115 VSIR -1.6561828 1.474093e-04 -2.5108181 7.589827e-08
68 5660 PSAP -0.5985553 5.893906e-10 -0.6485975 8.405687e-09
69 9806 SPOCK2 -0.7097012 6.273305e-07 -0.9577646 7.542568e-10
70 5328 PLAU -0.7200618 1.829326e-02 -1.7410365 9.083042e-12
71 132 ADK 0.8363206 4.083343e-02 1.2135609 3.886456e-03
72 23522 KAT6B 0.7101744 1.234099e-02 1.1743309 1.206557e-04
73 311 ANXA11 -0.8806356 7.784308e-05 -1.0562615 5.668088e-04
74 27063 ANKRD1 -1.4270897 1.153427e-03 -2.0426851 3.030782e-04
75 9124 PDLIM1 -0.4662416 3.761093e-03 -1.8420619 1.349910e-20
76 10580 SORBS1 0.7855235 2.886969e-02 1.2166623 7.183231e-04
77 83742 MARVELD1 -1.0286767 7.174480e-03 -3.0259238 1.527708e-05
78 84171 LOXL4 -1.0912243 4.996114e-03 -2.0187601 3.426042e-05
79 25911 DPCD 1.1067700 1.338041e-03 0.9704612 3.758772e-02
80 9118 INA 2.2944904 2.357074e-09 3.0493626 2.313032e-18
81 63877 FAM204A 1.2393674 4.485084e-03 1.1310294 4.145350e-02
82 9531 BAG3 -0.6233691 2.571072e-02 -0.8178849 2.026620e-03
83 64787 EPS8L2 -1.8935110 2.340663e-02 -2.5663294 1.858836e-03
84 6181 RPLP2 -0.7174814 2.210442e-12 -0.4741535 3.476979e-05
85 977 CD151 -0.8681080 7.588897e-08 -0.7357631 8.423508e-04
86 54472 TOLLIP -0.4982524 3.574692e-02 -0.6498546 2.745332e-03
87 1509 CTSD -1.3935398 4.407189e-11 -1.1722509 1.071834e-11
88 975 CD81 -0.6989561 1.534773e-10 -0.5289782 1.682520e-05
89 1200 TPP1 -0.8488296 3.009729e-09 -0.6858730 2.653874e-06
90 283298 OLFML1 -2.6003629 3.508751e-04 -4.4032135 1.447613e-11
91 8495 PPFIBP2 -1.9465948 4.024658e-03 -2.6782537 2.372853e-02
92 6157 RPL27A -0.4576164 1.138453e-05 -0.5739210 4.013843e-08
93 56672 AKIP1 -0.8686258 2.325365e-04 -1.0389619 1.177951e-02
94 100463486 <NA> -0.5670028 1.677433e-05 -0.7652167 1.678545e-11
95 10335 IRAG1 2.7682411 1.357927e-03 3.4438449 2.442476e-05
96 10418 SPON1 2.3280350 3.487032e-25 2.8478625 1.018164e-36
97 100126784 <NA> -1.1031581 3.597762e-02 -2.0769832 4.911540e-04
98 90139 TSPAN18 -1.2446392 8.470947e-03 -3.2097899 1.034229e-07
99 4192 MDK -0.7494072 3.003566e-10 -1.3108852 5.259237e-23
100 54972 TMEM132A -1.5776136 1.977545e-06 -1.2161426 1.815955e-02
101 1937 EEF1G -0.7085167 1.958882e-08 -0.3632931 2.369763e-02
102 6094 ROM1 -0.7308415 6.221272e-03 -0.7488741 2.219012e-02
103 51035 UBXN1 -0.6609201 4.991695e-03 -0.6196110 1.006333e-02
104 6520 SLC3A2 -0.9889574 1.618617e-12 -1.0115235 2.888269e-09
105 5920 PLAAT4 -0.8685048 5.434819e-07 -0.6796063 7.983631e-06
106 402 ARL2 -1.1010997 8.832002e-07 -0.7358065 4.016629e-03
107 4054 LTBP3 -0.9063847 9.822043e-05 -1.4519207 2.508272e-09
108 54961 SSH3 -0.9198075 9.399958e-03 -1.2848016 2.089242e-03
109 80194 TMEM134 -0.8107057 4.212660e-04 -0.8099869 2.372853e-02
110 1374 CPT1A -0.5600652 3.549433e-02 -0.8117826 7.467679e-04
111 55191 NADSYN1 -1.2628092 4.260543e-04 -1.0690123 3.470726e-02
112 6188 RPS3 -0.5220880 5.920265e-07 -0.4648421 2.598922e-05
113 2893 GRIA4 1.1706552 2.985746e-02 1.3518217 1.694079e-02
114 1410 CRYAB -0.9156951 1.652388e-06 -1.0653068 4.714061e-09
115 10525 HYOU1 -0.4883015 1.171868e-02 -0.4761770 4.158770e-02
116 403312 <NA> -3.0710157 2.124082e-02 -6.4439291 3.093675e-03
117 50937 CDON -1.2861130 1.410750e-02 -2.0978902 9.137794e-05
118 334 APLP2 -0.6864807 5.448047e-07 -0.5980690 3.633511e-08
119 84318 CCDC77 1.9559608 1.846516e-02 2.1511872 3.716361e-02
120 2597 GAPDH -0.6744001 8.190301e-11 -0.3867043 1.143118e-03
121 2 A2M 1.3474587 3.850375e-04 1.3629432 4.317881e-03
122 53919 SLCO1C1 2.1891277 4.202592e-03 3.5136960 9.991595e-10
123 51474 LIMA1 1.3616229 9.938361e-26 0.9004616 6.724941e-08
124 3164 NR4A1 1.9296629 8.141530e-03 2.9065407 2.530990e-06
125 84926 SPRYD3 -0.9005917 1.074380e-03 -1.1135619 3.033953e-04
126 3489 IGFBP6 -1.6158749 2.771846e-09 -2.8254651 3.159904e-19
127 967 CD63 -0.6319375 1.722591e-10 -0.3146512 4.581603e-02
128 4327 MMP19 -1.0299423 3.509051e-02 -2.3782134 1.632477e-03
129 1606 DGKA -0.8424901 7.612123e-03 -1.7099077 1.209739e-07
130 10956 OS9 -0.5445322 9.862403e-04 -0.3954939 2.732771e-02
131 4193 MDM2 -0.8867596 8.462040e-07 -0.7687663 7.925596e-06
132 10576 CCT2 0.6457436 2.425615e-05 0.4989460 2.578000e-02
133 6857 SYT1 1.6553199 6.653010e-16 1.7170863 1.103898e-11
134 1848 DUSP6 0.9381464 1.810766e-05 0.8078709 5.403907e-03
135 694 BTG1 0.8585758 2.682658e-04 1.5106789 1.368231e-10
136 7112 TMPO 1.1688164 9.282508e-03 1.2765529 1.019445e-02
137 429 ASCL1 -2.4723188 6.537856e-09 -1.5324763 1.191777e-03
138 51559 NT5DC3 -1.6587631 1.025955e-04 -2.5754704 4.174588e-07
139 5992 RFX4 0.8024374 3.748112e-02 1.1055461 5.672481e-03
140 9921 RNF10 -0.8939529 2.535337e-10 -0.5491389 2.306058e-03
141 100293704 <NA> 4.1563955 2.825374e-05 3.4350327 2.757241e-02
142 9612 NCOR2 -0.7602512 9.848057e-06 -0.5710013 1.021898e-03
143 100507206 LINC00943 6.2670067 1.041634e-02 7.3414130 9.659927e-04
144 219287 AMER2 2.2963669 6.465470e-03 2.5913085 3.626914e-03
145 2963 GTF2F2 -0.6050292 1.601933e-02 -0.8411555 3.360772e-03
146 1948 EFNB2 0.7101395 1.071572e-04 0.9160726 4.874069e-05
147 1284 COL4A2 -0.8486497 1.003297e-08 -0.8581541 8.485687e-09
148 4323 MMP14 -1.5278728 3.183308e-05 -0.9471221 2.920741e-02
149 26020 LRP10 -0.7125061 1.212563e-05 -0.5718418 9.659966e-04
150 5720 PSME1 -0.5474208 1.436507e-03 -0.5961568 2.539067e-03
151 9472 AKAP6 1.7299691 7.413449e-03 1.8351442 4.383108e-02
152 64067 NPAS3 -0.4257191 2.027830e-02 -0.5270289 8.920812e-03
153 6235 RPS29 -0.7419613 1.250840e-11 -0.3988568 3.375981e-03
154 54331 GNG2 1.7369460 1.854177e-06 2.3657889 2.163594e-12
155 10979 FERMT2 0.4448354 1.174916e-02 0.6132385 5.687382e-04
156 652 BMP4 3.4239509 2.447949e-02 3.9282995 1.794477e-02
157 1033 CDKN3 1.4117871 1.754875e-02 1.5761865 2.062667e-02
158 5015 OTX2 -3.8567439 1.522762e-04 -8.7825497 4.552874e-08
159 23002 DAAM1 0.5663210 1.303210e-02 0.6304340 1.524497e-02
160 6252 RTN1 1.2046072 1.295980e-10 2.0068226 1.885586e-24
161 57570 TRMT5 0.5383095 1.996713e-02 0.7275723 1.994641e-03
162 3306 HSPA2 1.0034327 1.000618e-02 1.4517179 5.152929e-04
163 87 ACTN1 -0.5041066 2.248568e-05 -0.6336172 6.001777e-06
164 10577 NPC2 -0.5348598 2.832452e-03 -0.6278263 2.755529e-04
165 83694 RPS6KL1 3.8201660 2.367539e-02 4.3708508 7.955020e-03
166 7043 TGFB3 -1.7755221 6.625823e-05 -1.6010358 6.498426e-04
167 9369 NRXN3 1.2076958 2.605854e-02 2.4114552 3.790622e-08
168 145508 CEP128 7.1295244 3.778787e-04 6.8727572 3.909650e-03
169 10516 FBLN5 1.3768596 1.003297e-08 2.7433114 3.778099e-29
170 4707 NDUFB1 -0.9902840 3.074268e-17 -0.3341957 3.882418e-02
171 90050 FAM181A 0.6619662 1.475180e-02 0.8686714 1.425327e-03
172 83982 IFI27L2 -0.7202890 1.524609e-04 -0.8509876 5.152929e-04
173 283596 SNHG10 -0.9927725 4.083343e-02 -1.3740020 3.135116e-02
174 64919 BCL11B 3.1371153 3.527181e-03 3.6254253 5.774574e-04
175 1778 DYNC1H1 -0.5485667 2.643472e-04 -0.6910382 7.226175e-04
176 7127 TNFAIP2 -1.2472245 1.314800e-07 -1.8952684 9.764034e-13
177 26153 KIF26A 1.5367998 1.627414e-03 2.5552542 4.666134e-12
178 7057 THBS1 1.1322840 4.725397e-02 2.3120156 1.526250e-07
179 388115 CCDC9B -0.9707164 9.390954e-03 -0.9800008 3.091238e-02
180 825 CAPN3 -2.5049072 6.996708e-03 -2.7096515 9.066743e-03
181 10169 SERF2 -0.5095081 4.008222e-04 -0.4437709 1.077343e-02
182 2628 GATM 1.0041797 1.277399e-04 2.1362447 7.146462e-22
183 9768 PCLAF 1.0548874 4.023695e-02 1.4751358 8.189029e-04
184 6176 RPLP1 -0.6184124 1.489460e-07 -0.4588419 2.538493e-05
185 5315 PKM -0.9462179 1.335801e-13 -1.1946909 1.515324e-20
186 3073 HEXA -0.6745240 4.093446e-04 -0.9780560 4.440033e-06
187 80381 CD276 -1.2080251 9.710680e-12 -0.7212907 4.517706e-04
188 4016 LOXL1 -1.1621947 9.591498e-12 -2.2308563 1.475835e-19
189 57611 ISLR2 -1.5610380 9.397460e-05 -1.5080340 9.766076e-04
190 1512 CTSH -0.9583259 6.302709e-03 -2.5144022 6.510032e-08
191 64782 AEN -0.7125988 1.841671e-02 -0.6638910 4.525681e-02
192 9055 PRC1 0.8420240 1.445306e-02 1.1261837 5.275301e-04
193 6627 SNRPA1 0.9282727 1.628912e-02 0.9524635 1.785120e-02
194 146330 FBXL16 1.2518787 7.372125e-03 1.1795037 3.470726e-02
195 1186 CLCN7 -0.9913242 1.581963e-04 -0.9764001 1.804932e-03
196 100507303 <NA> -1.6805316 4.220443e-15 -0.5121858 4.520012e-02
197 9235 IL32 -0.6818497 1.842470e-02 -1.5879467 1.420426e-07
198 84662 GLIS2 -0.8247250 2.320839e-03 -0.7922237 2.174280e-02
199 2903 GRIN2A 2.2027307 1.186896e-03 2.3038080 3.160988e-03
200 8303 SNN 0.4455792 4.644851e-02 0.5221263 4.096260e-02
201 51704 GPRC5B -0.4680241 1.239776e-02 -0.5989051 3.145272e-03
202 388228 SBK1 0.6882155 3.388523e-02 1.3196507 1.756807e-05
203 25970 SH2B1 -0.5829693 2.921922e-02 -0.6386420 4.342193e-02
204 4502 MT2A -0.8684887 2.126854e-13 -0.7408360 1.655015e-07
205 6236 RRAD -1.2954374 5.170409e-08 -1.6201892 2.264646e-10
206 8824 CES2 -0.6597458 2.165159e-04 -0.6495872 2.871545e-03
207 5699 PSMB10 -0.9433930 1.457500e-03 -0.9026673 1.290766e-02
208 6560 SLC12A4 -1.2108080 1.003837e-04 -1.1917565 2.167243e-05
209 999 CDH1 -2.6912992 3.675478e-03 -2.7009634 9.781015e-03
210 794 CALB2 1.4563435 2.088537e-14 2.7326036 4.233752e-48
211 497190 CLEC18B;CLEC18C -1.7466351 1.406097e-05 -2.3578587 1.134816e-09
212 57687 VAT1L 1.0462980 5.389711e-10 1.3416085 9.018065e-15
213 55839 CENPN 1.9090533 4.758100e-04 1.8937478 3.155924e-03
214 80790 CMIP 0.6259151 1.220729e-02 1.0032746 8.047882e-05
215 23199 GSE1 0.6500420 3.495892e-03 0.8982309 9.960357e-04
216 8140 SLC7A5 -1.0824671 7.858961e-22 -2.2452474 1.547199e-54
217 6137 RPL13 -0.7156228 2.316918e-06 -0.3737988 3.926732e-02
218 10381 TUBB3 0.5010398 2.416989e-03 0.9889765 1.199731e-10
219 79007 DBNDD1 -1.2128263 4.882144e-04 -1.3401715 1.812461e-04
220 11337 GABARAP -0.8348070 7.715736e-08 -0.3634791 4.902417e-02
221 5187 PER1 0.7651457 3.149675e-02 1.7124505 6.582345e-10
222 4628 MYH10 0.4612805 1.372416e-02 0.4664788 4.737911e-02
223 9423 NTN1 -0.8132813 1.369377e-05 -0.5323178 1.166804e-02
224 5376 PMP22 0.5824850 4.264601e-03 0.5750672 1.361909e-02
225 201161 CENPV 1.2126377 2.236816e-07 2.0594058 2.654752e-23
226 51655 RASD1 0.9198690 3.515780e-03 1.5883485 1.716524e-08
227 146691 TOM1L2 -0.6079747 6.706658e-04 -0.6006084 3.145272e-03
228 8851 CDK5R1 0.7194148 3.138099e-02 1.4191421 2.630099e-07
229 124842 TMEM132E -1.1219998 4.794497e-05 -0.9900478 3.356139e-03
230 7153 TOP2A 0.6511570 1.137433e-02 1.1935666 2.066820e-08
231 60681 FKBP10 -0.4783603 9.282508e-03 -0.8373423 9.942255e-06
232 4669 NAGLU -1.1848892 2.049626e-05 -0.7648877 2.447416e-02
233 10493 VAT1 -0.4830937 1.805737e-02 -0.6498699 1.231201e-03
234 8153 RND2 -0.9388098 7.327209e-04 -0.7153644 3.690794e-02
235 10900 RUNDC3A 3.4232521 1.037063e-03 4.2168733 2.487972e-06
236 51629 SLC25A39 -0.7710945 5.536538e-04 -0.6620026 1.644075e-02
237 2896 GRN -0.8731183 6.200187e-10 -1.0363383 3.516910e-10
238 23131 GPATCH8 0.9859784 8.951123e-03 1.0218764 2.447416e-02
239 2535 FZD2 -0.8609953 5.812486e-03 -0.7855913 2.362308e-02
240 2670 GFAP -0.8880523 2.924309e-14 -0.6316674 1.689940e-12
241 4836 NMT1 -1.1723199 1.030154e-14 -0.6540254 3.325038e-04
242 113026 PLCD3 -0.6555781 9.747615e-04 -1.0411326 1.978628e-07
243 4804 NGFR -1.9056027 1.518377e-04 -2.0854285 5.774088e-05
244 9902 MRC2 -0.4766944 2.269338e-04 -0.8065596 5.349633e-08
245 2232 FDXR -1.1088438 2.249946e-04 -0.9142345 7.479277e-03
246 3021 H3-3B 0.4388141 1.639482e-02 0.5520628 2.606542e-03
247 91107 TRIM47 -0.5611067 3.970094e-02 -1.1071090 2.279490e-04
248 6427 SRSF2 0.6480996 1.038843e-03 0.7560787 1.133735e-03
249 57690 TNRC6C 0.9229884 2.032680e-03 1.2408552 4.864067e-05
250 7077 TIMP2 -0.5543884 1.204285e-05 -0.5314700 3.524744e-03
251 3959 LGALS3BP -1.0898674 5.878910e-11 -1.0120031 6.988221e-10
252 2548 GAA -1.1102211 2.941173e-04 -1.1230197 1.954423e-03
253 57674 RNF213 -0.8381919 1.004688e-03 -0.8197228 9.066743e-03
254 284184 NDUFAF8 -0.7896434 2.779541e-07 -0.4036235 1.838629e-02
255 124565 SLC38A10 -0.9287498 1.547999e-06 -0.7670923 8.223759e-03
256 5034 P4HB -0.4904015 4.422442e-04 -0.4531657 1.332349e-02
257 23136 EPB41L3 0.7952735 1.970178e-02 1.2231705 5.820257e-05
258 361 AQP4 1.9744058 1.884676e-21 2.4605834 5.192154e-20
259 8715 NOL4 2.5588222 5.457858e-05 2.3582394 4.119081e-03
260 56853 CELF4 2.8209731 1.393291e-02 3.6577947 1.803873e-04
261 5596 MAPK4 2.1524133 1.618136e-05 3.1760688 8.647871e-15
262 51320 MEX3C 0.7125434 1.606521e-03 0.7097423 1.580893e-02
263 6925 TCF4 0.5133689 9.417442e-03 0.6808140 3.298958e-03
264 51046 ST8SIA3 1.7149492 5.575706e-03 2.0068729 1.401190e-03
265 115701 ALPK2 -0.3734904 1.009453e-02 -0.9164225 3.602790e-06
266 90701 SEC11C 0.6300709 1.026419e-02 0.6954960 1.566203e-02
267 22850 ADNP2 0.9290115 3.211088e-02 1.0547023 3.869495e-02
268 10272 FSTL3 -1.9231970 1.242140e-08 -2.4261528 9.126936e-09
269 2879 GPX4 -0.8032023 9.131109e-05 -0.5574165 7.786075e-03
270 60680 CELF5 1.6306300 1.111506e-04 2.8128217 2.643450e-18
271 10362 HMG20B -0.5795260 2.201606e-02 -0.6495918 3.781472e-02
272 1938 EEF2 -0.7481511 8.743561e-11 -0.6570017 1.299284e-06
273 80700 UBXN6 -1.3411933 3.086274e-07 -0.5621434 3.470726e-02
274 25873 RPL36 -1.1973264 1.077139e-23 -0.4972307 9.698640e-04
275 126328 NDUFA11 -0.8775321 1.958882e-08 -0.4355587 4.136950e-02
276 3383 ICAM1 -0.5475674 3.043923e-02 -0.9747244 6.115328e-03
277 7297 TYK2 -0.8441545 3.482509e-03 -0.8571501 8.414839e-03
278 1995 ELAVL3 1.5915190 4.203001e-14 2.9384178 2.628971e-63
279 51398 WDR83OS -0.6712717 4.639231e-06 -0.5014992 7.895365e-03
280 4784 NFIX -0.5865548 1.171868e-02 -0.6984317 7.419264e-03
281 9592 IER2 0.7818117 1.312546e-03 0.8378179 1.025390e-02
282 23025 UNC13A 1.9715346 3.989972e-02 2.9582727 5.511003e-04
283 54858 PGPEP1 -1.1903414 3.073928e-11 -1.2047971 6.065331e-09
284 1463 NCAN 0.5372292 5.508163e-04 1.3587582 3.010784e-20
285 57616 TSHZ3 2.1966430 1.003716e-03 2.7114870 2.719685e-05
286 1054 CEBPG 2.4854524 7.843094e-11 1.8922275 3.095694e-04
287 9710 GARRE1 -0.6555832 1.456301e-02 -0.8321661 5.924183e-03
288 7392 USF2 -0.9962607 3.825282e-06 -0.5331062 3.804954e-02
289 826 CAPNS1 -0.9240183 7.655060e-10 -0.6648593 1.818685e-04
290 7538 ZFP36 2.0965628 8.061713e-05 2.1072748 1.377638e-03
291 645 BLVRB -1.0425590 1.061855e-03 -1.1143163 3.961323e-04
292 602 BCL3 -0.6415942 1.445306e-02 -1.5072211 5.265819e-09
293 27113 BBC3 -1.3396761 1.444217e-14 -1.1519733 1.140833e-09
294 8605 PLA2G4C -1.3646974 3.798788e-02 -1.6907504 4.257213e-02
295 10945 KDELR1 -0.3259247 3.425822e-02 -0.5403373 4.295628e-03
296 6141 RPL18 -0.4015145 9.783400e-04 -0.3351432 1.357813e-02
297 51171 HSD17B14 -2.4475629 7.587371e-06 -3.0654508 8.323988e-07
298 4924 NUCB1 -0.8898897 4.396609e-08 -0.5295417 6.115328e-03
299 581 BAX -1.5018632 1.472426e-13 -0.7553891 7.965274e-06
300 2512 FTL -0.5033572 2.071844e-03 -0.6857683 6.851876e-09
301 2217 FCGRT -0.8874348 6.210984e-04 -1.0540916 1.737421e-03
302 6237 RRAS -1.0583985 1.814848e-05 -1.7603488 2.465258e-06
303 2109 ETFB -0.9172335 8.043166e-06 -0.5426740 2.598169e-02
304 147807 ZNF524 -1.7047506 3.039626e-02 -1.7491326 4.941566e-02
305 101060391 <NA> 2.4715761 1.427430e-02 3.7036718 2.621680e-06
306 6664 SOX11 0.6311761 3.548487e-05 1.8473670 4.632773e-31
307 57498 KIDINS220 0.6079342 4.524642e-03 0.6657516 7.176441e-03
308 6382 SDC1 -1.3382067 3.794705e-03 -1.7751396 2.960107e-03
309 5500 PPP1CB -0.3566350 1.764587e-02 -1.0157787 1.401469e-10
310 6432 SRSF7 0.9134427 4.093446e-04 0.8839854 2.387854e-03
311 9378 NRXN1 3.2466511 1.225951e-04 4.3584971 4.110696e-10
312 55704 CCDC88A 1.3788688 2.389737e-20 1.1202549 2.653548e-08
313 51255 RNF181 -0.6098092 1.128883e-03 -0.7259563 8.727483e-04
314 51652 CHMP3 -1.0700679 1.737304e-05 -0.9002936 1.351186e-03
315 400986 ANKRD36C 1.0433345 1.065900e-03 0.9147547 2.388319e-02
316 57730 ANKRD36B 0.9923870 2.687596e-04 0.8825329 2.092788e-02
317 9486 CHST10 -0.9085586 1.473834e-07 -0.9388194 3.342742e-08
318 84417 ECRG4 1.0156780 2.552547e-03 1.1657628 1.055937e-03
319 3625 INHBB -1.1562493 1.364735e-05 -1.6228571 1.002300e-04
320 2863 GPR39 -1.6579966 2.066066e-03 -2.2619382 1.890523e-04
321 4249 MGAT5 0.7354107 1.063177e-02 0.9656419 1.185644e-03
322 3800 KIF5C 0.4556294 1.763758e-02 0.7199533 4.277784e-04
323 114793 FMNL2 -0.7987794 1.171565e-05 -0.8717975 2.719685e-05
324 56475 RPRM 2.6393333 2.692385e-02 3.7420358 3.679967e-04
325 90 ACVR1 1.2420487 1.899938e-02 1.5871805 3.023614e-03
326 220988 HNRNPA3 0.6389881 5.964460e-04 0.6473359 2.815547e-03
327 91404 SESTD1 1.6600210 1.111506e-04 1.5351811 9.066743e-03
328 51602 NOP58 1.2025070 2.864822e-06 1.0514439 1.143118e-03
329 8828 NRP2 -0.5317226 2.425229e-03 -0.9764990 4.361339e-09
330 8609 KLF7 0.9936242 1.325519e-06 0.9866636 4.517706e-04
331 2066 ERBB4 2.2396353 4.100469e-04 2.8991771 1.167719e-07
332 79582 SPAG16 0.6190746 4.700875e-03 0.7705482 1.955819e-03
333 6168 RPL37A -0.9396317 8.680184e-19 -0.3840962 4.163477e-03
334 3488 IGFBP5 -1.4896444 8.072817e-13 -2.6454977 5.652670e-27
335 5798 PTPRN 6.8726501 9.083539e-04 7.7871502 3.476979e-05
336 79586 CHPF -1.0246442 5.003934e-08 -0.5407254 4.362068e-02
337 7857 SCG2 2.6021431 7.706302e-10 3.7148838 2.193291e-25
338 5270 SERPINE2 -1.0018258 8.026516e-08 -1.4460396 4.552874e-08
339 151473 SLC16A14 6.8885107 9.039198e-04 7.1940831 1.593263e-03
340 5757 PTMA 0.7428128 1.652843e-04 0.7707692 1.442434e-04
341 3635 INPP5D -2.2678809 5.208667e-03 -2.8373930 6.669893e-04
342 2637 GBX2 -2.7097991 4.293880e-03 -6.3088695 1.342612e-04
343 2817 GPC1 -0.7105645 1.457500e-03 -0.7075240 3.028675e-03
344 83959 SLC4A11 -1.8193726 1.233695e-02 -2.3851510 4.644121e-02
345 57506 MAVS -0.9340439 4.124753e-05 -0.8765917 2.085698e-04
346 6238 RRBP1 -0.8779930 2.020395e-05 -0.9180331 2.592803e-04
347 2937 GSS -0.7534159 8.576030e-03 -1.0892960 8.111724e-04
348 4826 NNAT 3.1581106 1.194518e-16 5.3255153 1.206740e-19
349 84969 TOX2 -1.1548905 6.373651e-04 -1.3513027 9.689222e-04
350 51604 PIGT -0.5082443 3.110183e-02 -0.7031226 6.982086e-03
351 11065 UBE2C 1.3272127 3.482509e-03 1.7874888 3.257545e-04
352 5740 PTGIS -2.7710800 1.890294e-02 -7.0784709 9.728671e-05
353 100462983 <NA> -0.9910046 3.609496e-03 -1.1828045 1.656836e-03
354 56937 PMEPA1 -0.5320660 3.022419e-03 -0.9454583 1.836351e-07
355 3911 LAMA5 -0.6144204 8.420533e-03 -1.2663307 1.409309e-08
356 6227 RPS21 -0.5685228 1.775938e-06 -0.4278776 1.600810e-03
357 79144 PPDPF -0.6761500 2.618381e-06 -0.4118019 1.890401e-02
358 4661 MYT1 5.8098946 7.668167e-03 7.5689872 1.158351e-05
359 140578 CHODL 0.8540394 2.124082e-02 1.2471015 1.943344e-03
360 10600 USP16 1.2808729 6.492063e-09 0.7838947 1.566203e-02
361 56245 C21orf62 0.8845232 6.769030e-04 0.9911059 1.890523e-04
362 1827 RCAN1 1.6077177 4.528098e-32 1.7948495 5.211876e-32
363 5211 PFKL -1.0086018 5.135812e-06 -0.8106155 1.782628e-03
364 80781 COL18A1 -0.6519162 3.489677e-03 -1.4193803 4.072636e-11
365 1291 COL6A1 -0.7424651 2.474967e-07 -0.6750073 2.264092e-05
366 1292 COL6A2 -1.3257081 8.242001e-04 -1.4716832 1.370357e-03
367 114044 MCM3AP-AS1 4.8345090 2.117025e-02 5.1403831 2.858695e-02
368 9993 DGCR2 -0.7484963 6.685442e-07 -0.4764508 1.488234e-02
369 652968 CASTOR1 -0.9054170 9.394588e-03 -1.0721755 5.891976e-03
370 6948 TCN2 -1.3809025 3.774116e-04 -1.2593961 5.521371e-03
371 8542 APOL1 -2.4446249 3.469519e-03 -2.7218890 4.159163e-04
372 4627 MYH9 -0.3778974 4.183716e-03 -0.6971789 5.066593e-05
373 29775 CARD10 -0.9491607 6.527177e-03 -1.3116225 2.320783e-02
374 27350 APOBEC3C -1.9712267 5.060477e-06 -2.6040539 1.588902e-06
375 6122 RPL3 -0.4053336 3.227527e-04 -0.4766353 2.816087e-04
376 468 ATF4 0.7251146 2.816812e-04 0.6186308 1.994641e-03
377 55964 SEPTIN3 0.6445300 1.469193e-04 0.9447643 4.142801e-08
378 112885 PHF21B 1.5458340 4.350773e-03 2.2259992 8.294663e-06
379 55007 FAM118A -1.0390227 1.014990e-02 -1.3407462 3.962503e-02
380 375323 LHFPL4 3.2914513 1.100144e-02 5.0276729 2.919134e-09
381 7508 XPC -0.7197664 7.140592e-03 -0.7392739 2.928362e-02
382 9975 NR1D2 -1.2999325 5.263202e-03 -1.1804262 2.990120e-02
383 7068 THRB 5.0064347 2.270573e-03 6.0866477 1.309467e-05
384 116135 LRRC3B 1.1465580 1.021039e-03 1.4798345 1.818685e-04
385 27303 RBMS3 -2.0755081 7.668167e-03 -1.9865338 4.317191e-02
386 91392 ZNF502 2.1324500 4.337593e-02 2.7812227 3.342794e-03
387 10675 CSPG5 0.8353689 1.601933e-02 1.5784377 1.185997e-06
388 4134 MAP4 -0.3390575 4.375203e-03 -0.2940204 3.587645e-02
389 2771 GNAI2 -0.4636031 8.569642e-05 -0.4944595 2.408297e-05
390 7869 SEMA3B -1.6564932 3.862308e-08 -1.4513602 1.745701e-04
391 440957 SMIM4 -0.5958505 5.117766e-04 -0.5449620 2.265123e-03
392 7086 TKT -0.9017131 4.610312e-12 -1.1391536 8.482534e-15
393 2317 FLNB -0.6845340 3.984342e-02 -1.1857779 2.745332e-03
394 11170 FAM107A 1.1009337 2.709190e-03 2.4321216 5.947464e-17
395 132204 SYNPR 3.1683107 7.361290e-13 3.1351647 8.666835e-12
396 9223 MAGI1 0.4886117 1.660351e-02 0.6969315 2.438204e-02
397 56650 CLDND1 1.0712947 1.990708e-04 0.8798926 3.725134e-02
398 961 CD47 0.8238312 1.930611e-05 0.8183946 1.397105e-04
399 55081 IFT57 1.0869656 3.821786e-05 0.9825755 9.306479e-03
400 29114 TAGLN3 0.8215485 3.700229e-03 1.9259335 6.780918e-15
401 79858 NEK11 1.3326372 1.996092e-02 1.4479830 3.333364e-02
402 7018 TF -3.3232865 2.405283e-35 -2.6389352 1.155659e-27
403 5806 PTX3 1.8501347 1.754627e-11 1.4100724 2.477616e-03
404 6657 SOX2 -0.5220920 5.263072e-05 -0.4677946 1.999204e-02
405 84002 B3GNT5 3.1685177 1.047117e-04 2.9678226 4.211058e-03
406 6480 ST6GAL1 5.2799963 3.583567e-02 6.4016047 2.265123e-03
407 6165 RPL35A -0.3520399 1.015559e-02 -0.3375458 4.897053e-02
408 53834 FGFRL1 -0.8610945 4.109856e-04 -1.0660824 1.138515e-04
409 10417 SPON2 -2.3182470 1.543773e-08 -4.2936110 9.946733e-14
410 285489 DOK7 -1.6848822 2.773213e-02 -3.3149758 2.597013e-03
411 4043 LRPAP1 -0.4776386 1.800286e-03 -0.6202236 2.425953e-04
412 27065 NSG1 1.4547421 1.538258e-03 2.5616306 3.941577e-12
413 57537 SORCS2 -0.7963868 4.223483e-02 -1.6908825 1.012552e-04
414 8532 CPZ -0.7817281 5.583150e-04 -1.1441708 6.434948e-06
415 79730 NSUN7 3.1925786 8.091324e-03 3.7129693 1.177388e-03
416 9662 CEP135 2.3716028 3.901395e-03 2.3674758 2.085977e-02
417 57482 CRACD 1.4048826 1.534526e-03 1.3386405 3.473845e-02
418 2926 GRSF1 0.6163410 3.973976e-02 0.7575895 1.878569e-02
419 9508 ADAMTS3 1.2487115 3.219427e-02 1.8042029 5.566136e-03
420 9987 HNRNPDL 0.7576618 1.222739e-09 0.8450377 5.997885e-11
421 8404 SPARCL1 3.4421811 1.890294e-02 5.6833297 1.581164e-09
422 6164 RPL34 -0.2818415 9.384072e-03 -0.4235520 6.916522e-04
423 5188 GATB -0.6434372 8.809308e-03 -1.0747561 3.095694e-04
424 84057 MND1 6.3595666 8.312957e-04 5.7683103 2.228943e-02
425 2982 GUCY1A1 -2.5615469 3.073185e-02 -2.4540368 3.596483e-02
426 3148 HMGB2 1.5486097 4.131207e-07 1.7903237 1.320091e-06
427 79682 CENPU 1.7489542 2.358664e-02 1.9973075 1.599367e-02
428 134121 C5orf49 1.2796871 2.310883e-02 1.6368077 1.552128e-03
429 100505738 MIR4458HG -0.6829890 3.864705e-04 -0.9980030 1.665149e-07
430 6167 RPL37 -0.9213204 1.407983e-12 -0.3598677 3.596070e-02
431 3157 HMGCS1 0.8161423 1.619626e-04 1.1873172 5.061542e-09
432 5144 PDE4D 3.0150852 1.278346e-02 3.9688351 8.997510e-05
433 3796 KIF2A 0.7119465 2.298569e-02 0.9533351 1.000226e-02
434 4131 MAP1B 0.6409932 1.392403e-11 0.4285036 8.629840e-03
435 8507 ENC1 0.7105075 8.417686e-03 1.0075880 4.079814e-05
436 3156 HMGCR 1.1901546 1.788473e-06 1.0787649 2.908894e-04
437 7060 THBS4 1.6625515 3.700229e-03 1.9177103 1.879318e-03
438 1070 CETN3 -0.7720163 3.553947e-03 -0.5855236 4.941566e-02
439 7025 NR2F1 -0.5331827 4.331730e-03 -0.7465323 1.383557e-02
440 1946 EFNA5 1.7976054 3.221947e-03 2.3055068 1.364406e-04
441 324 APC 0.8236851 2.127409e-03 0.9432079 9.689222e-04
442 441108 <NA> -0.7490272 3.007057e-02 -1.0915868 2.447416e-02
443 23338 JADE2 -2.1536622 1.849520e-03 -1.8847536 1.607384e-02
444 9879 DDX46 0.4316611 3.755352e-02 0.6885911 3.449829e-03
445 9547 CXCL14 -1.1592910 2.264139e-15 -2.3882954 1.748834e-08
446 6695 SPOCK1 -1.4323253 2.985063e-06 -1.7771606 1.564048e-08
447 1958 EGR1 2.2574398 4.100469e-04 2.4815105 1.195813e-03
448 1839 HBEGF 0.7060774 3.219126e-02 1.2849143 2.137062e-04
449 2246 FGF1 0.6143881 3.167065e-02 1.0136959 1.629543e-03
450 11346 SYNPO -1.2061350 4.329632e-09 -1.2058775 8.530738e-06
451 10318 TNIP1 -1.1392066 3.351148e-08 -1.2069096 1.313948e-05
452 6678 SPARC -0.3085683 4.644851e-02 -0.5909387 2.442476e-05
453 345630 FBLL1 1.0156913 3.720260e-02 1.4519761 2.339917e-03
454 64901 RANBP17 7.3193637 1.025955e-04 6.1720844 4.190071e-02
455 1843 DUSP1 2.3289940 1.390953e-03 2.5480526 3.133156e-03
456 8992 ATP6V0E1 -0.3468481 3.024070e-02 -0.6275270 3.033400e-04
457 51617 NSG2 1.7956096 9.711090e-03 4.1012064 1.097758e-18
458 10814 CPLX2 1.6186017 1.043166e-03 3.4959251 7.251559e-29
459 11282 MGAT4B -0.7603028 1.195494e-05 -0.7867426 1.078348e-04
460 54898 ELOVL2 0.7891252 5.703222e-03 1.1113025 3.876063e-04
461 84062 DTNBP1 -1.1467664 2.702368e-05 -0.7257966 4.145350e-02
462 3400 ID4 1.3146187 3.369625e-14 0.9409950 7.382052e-06
463 10590 SCGN 2.4250008 7.037777e-26 3.1688152 5.652459e-49
464 100129195 ZSCAN16-AS1 -0.9919782 2.318761e-03 -0.9874230 2.372853e-02
465 10537 UBD -1.3605891 1.568922e-04 -3.7566251 2.600006e-15
466 3105 HLA-A -0.7721547 2.367539e-02 -1.1999501 6.081451e-04
467 3106 HLA-B -0.7012609 3.856387e-04 -0.9622349 3.891084e-06
468 6892 TAPBP -1.1018289 1.253132e-09 -0.8933290 2.206121e-07
469 221491 SMIM29 -0.9218884 2.768231e-04 -0.8155327 1.821625e-02
470 6428 SRSF3 0.5815088 1.925323e-03 0.5327334 3.333364e-02
471 1026 CDKN1A -0.7445167 2.958591e-09 -1.0389839 9.994481e-13
472 221477 C6orf89 -0.4383175 3.119456e-02 -0.6176692 2.968614e-03
473 4188 MDFI -1.7036101 3.845899e-07 -1.4214050 5.451974e-03
474 6722 SRF 0.4945680 3.925916e-02 0.6543793 2.320783e-02
475 5429 POLH -0.8572630 4.996114e-03 -0.8523754 3.111382e-02
476 441151 TMEM151B 2.4714638 2.246212e-02 3.8518943 1.661677e-06
477 10492 SYNCRIP 0.6456309 1.146635e-04 0.6673359 5.668088e-04
478 10690 FUT9 1.6135560 4.496886e-02 2.0335869 1.478942e-02
479 100133941 CD24 0.7736300 4.061450e-04 1.0346292 5.668484e-06
480 2534 FYN 0.5662551 2.445531e-03 0.5032670 3.061403e-02
481 2697 GJA1 0.7769008 2.142055e-09 0.6745041 2.252541e-07
482 1490 CCN2 1.7576486 4.740599e-38 1.4848577 2.026620e-03
483 51696 HECA 2.2213221 8.253492e-03 2.0533598 4.927610e-02
484 168090 C6orf118 6.5370887 9.912320e-06 7.1098054 7.340385e-07
485 51660 MPC1 1.3771688 1.179438e-16 0.8873961 2.265123e-03
486 392617 ELFN1 -0.7723583 1.361221e-02 -0.9297286 1.119240e-02
487 29886 SNX8 -0.8195445 1.012372e-03 -0.7999480 5.254358e-03
488 2115 ETV1 1.6879494 7.614340e-28 1.6873638 3.754955e-16
489 8701 DNAH11 6.6067617 3.610518e-03 7.3324766 1.005705e-03
490 4852 NPY 1.3065693 2.221224e-06 2.3309502 5.128796e-28
491 441204 <NA> -1.5495794 4.241548e-02 -3.8773492 1.752408e-04
492 79017 GGCT 1.2005300 5.250533e-06 0.9920255 1.564745e-02
493 64759 TNS3 -0.6747864 1.569101e-02 -1.2948669 1.039388e-05
494 7378 UPP1 -0.6847359 5.323075e-03 -0.9439945 8.024406e-04
495 64409 GALNT17 -1.0330728 5.765903e-06 -1.9242295 7.281613e-03
496 11257 <NA> -0.8846712 3.157582e-02 -1.2390700 1.280812e-02
497 5244 ABCB4 -0.8029967 1.000909e-02 -1.7639557 1.443107e-08
498 1750 DLX6 1.6876904 1.074380e-03 2.9617545 2.577095e-16
499 1749 DLX5 2.1403466 3.330027e-05 3.6286422 3.355781e-22
500 25798 BRI3 -0.5697038 2.528938e-05 -0.4978698 1.195813e-03
501 245812 CNPY4 -0.6882340 3.677197e-02 -1.0333447 1.364081e-02
502 7205 TRIP6 -0.5610654 1.128883e-03 -0.4880182 1.476705e-02
503 5054 SERPINE1 2.8428159 3.704122e-19 2.6220037 4.061962e-14
504 2861 GPR37 1.9524966 2.425447e-04 2.6224191 2.354399e-09
505 9296 ATP6V1F -0.6297690 3.603297e-03 -0.5086225 2.619702e-02
506 4232 MEST 1.0997440 9.949038e-06 1.8222392 1.563627e-15
507 26958 COPG2 2.0279355 3.071839e-08 2.3778189 7.107555e-09
508 155066 ATP6V0E2 -0.9387413 4.901488e-10 -0.5342727 4.860574e-04
509 54480 CHPF2 -0.6816237 3.425822e-02 -0.8327015 2.138184e-02
510 349136 WDR86 -1.1448503 1.561161e-09 -1.1317281 1.661758e-04
511 79660 PPP1R3B 1.4515542 1.555100e-03 1.8445544 5.408729e-05
512 10395 DLC1 1.8666561 2.280812e-16 1.7153743 5.997885e-11
513 4747 NEFL -1.9499689 7.411800e-07 -2.8381102 5.437741e-10
514 1808 DPYSL2 0.7938179 4.526794e-10 0.9699894 2.989223e-14
515 1191 CLU -0.3536011 8.258154e-03 -0.6740472 2.714996e-07
516 80223 RAB11FIP1 -1.7252299 1.777853e-05 -2.8727814 3.109746e-05
517 286183 NKAIN3 -0.9095352 1.619177e-06 -1.0933370 2.559890e-05
518 8836 GGH -0.4446193 4.961455e-02 -1.4253748 3.883817e-10
519 5569 PKIA 1.2302738 4.545022e-03 1.2067039 2.921959e-02
520 11075 STMN2 5.0178692 4.039958e-03 6.1978578 3.966498e-04
521 862 RUNX1T1 2.3534165 9.067719e-06 3.3089522 1.119515e-13
522 7381 UQCRB -0.3130180 4.232915e-02 -0.3555414 1.242384e-02
523 79870 BAALC 0.5156894 2.569769e-04 0.4336691 1.656836e-03
524 4062 LY6H 2.3281384 9.327216e-04 3.5223270 2.754717e-10
525 1936 EEF1D -1.1117231 2.704108e-05 -0.6600540 9.022899e-03
526 113655 MFSD3 -1.2694565 1.886682e-04 -0.9020509 4.446365e-02
527 6132 RPL8 -0.7008929 8.295695e-13 -0.5470781 4.292663e-06
528 6595 SMARCA2 -0.5005963 2.128126e-02 -0.7168505 1.237660e-03
529 11168 PSIP1 1.0311192 7.655060e-10 0.4980641 4.166996e-02
530 92949 ADAMTSL1 -1.5747186 1.178586e-02 -2.3156612 5.553919e-03
531 10210 TOPORS 0.8612838 2.257219e-02 0.8828085 4.667604e-02
532 2683 B4GALT1 -0.7110255 3.176405e-03 -1.5339468 1.558087e-05
533 7094 TLN1 -0.5595991 1.057199e-02 -0.6843306 1.552128e-03
534 389765 <NA> 5.4167190 2.133071e-02 5.9018589 1.550150e-02
535 8013 NR4A3 1.6926032 5.653334e-03 1.7210198 1.734746e-02
536 19 ABCA1 -0.3545432 4.929436e-02 -1.1633278 8.704903e-14
537 5998 RGS3 -0.4205005 4.337593e-02 -0.7371648 1.573076e-03
538 3371 TNC 1.4887713 2.568738e-02 1.9836646 2.549804e-03
539 5069 PAPPA -2.2794329 9.571169e-03 -3.7850262 2.918134e-03
540 1620 BRINP1 2.2480329 1.627583e-07 3.0111835 2.505557e-15
541 6136 RPL12 -0.3982599 3.179378e-03 -0.6014169 4.943460e-06
542 64855 NIBAN2 -0.5689618 2.714784e-02 -0.7917959 7.940203e-03
543 10044 SH2D3C 2.6863991 2.729971e-02 3.4168381 1.952613e-03
544 114789 SLC25A25 0.9565245 7.851947e-04 0.9885164 1.600810e-03
545 5524 PTPA -0.6516898 2.894609e-03 -0.8141781 6.648189e-04
546 100506119 LINC01503 -1.1269390 1.317657e-02 -1.7671980 4.880285e-04
547 1289 COL5A1 -1.2112262 3.804089e-03 -2.0207849 6.255122e-07
548 4851 NOTCH1 -0.5435727 3.013578e-02 -0.7688592 5.924183e-03
549 4558 <NA> -0.4390864 1.128883e-03 -0.3839364 9.338884e-03
550 4577 <NA> -1.3386473 7.087322e-15 -1.2498384 5.280320e-11
551 4567 <NA> -1.4240347 4.078910e-27 -0.5383313 1.321248e-03
552 4578 <NA> -0.6840371 3.484701e-02 -1.0416733 9.766076e-04
553 1654 DDX3X 1.1414177 1.042344e-06 0.8214014 6.765529e-03
554 4129 MAOB 0.9057778 1.524609e-04 1.4594028 1.659212e-12
555 57477 SHROOM4 -1.3916576 4.680550e-02 -2.0099208 2.076708e-02
556 170685 NUDT10 1.4053316 2.996299e-02 1.9828991 1.181037e-03
557 28986 MAGEH1 1.2557662 4.701573e-03 1.7791003 4.271693e-05
558 4675 NAP1L3 1.7653736 4.230242e-04 1.8870734 2.265123e-03
559 6173 RPL36A -0.7106463 7.262684e-03 -0.8018508 8.672044e-03
560 11013 TMSB15A 1.5281486 3.468449e-05 2.7261900 1.113867e-23
561 55859 BEX1 0.3781885 1.716155e-02 0.3878479 2.939565e-02
562 51186 TCEAL9 -0.3892609 3.662576e-03 -0.3464859 1.041252e-02
563 5063 PAK3 1.4575697 3.804089e-03 1.5492220 1.059705e-02
564 1641 DCX 1.7378594 1.806928e-19 3.0735949 4.554326e-68
565 3597 IL13RA1 -0.6852056 4.146922e-02 -1.1106445 1.936129e-02
566 6170 RPL39 -0.4891487 3.418020e-05 -0.4737819 1.080627e-03
567 3897 L1CAM 0.9307928 4.521374e-02 1.3151852 4.894149e-03
568 2316 FLNA -0.6322225 4.888813e-07 -0.5283306 1.251199e-03
569 8270 LAGE3 -0.5948944 2.044780e-02 -0.8901983 2.894585e-03
570 2539 G6PD -0.5669937 4.023695e-02 -0.8645735 7.784778e-03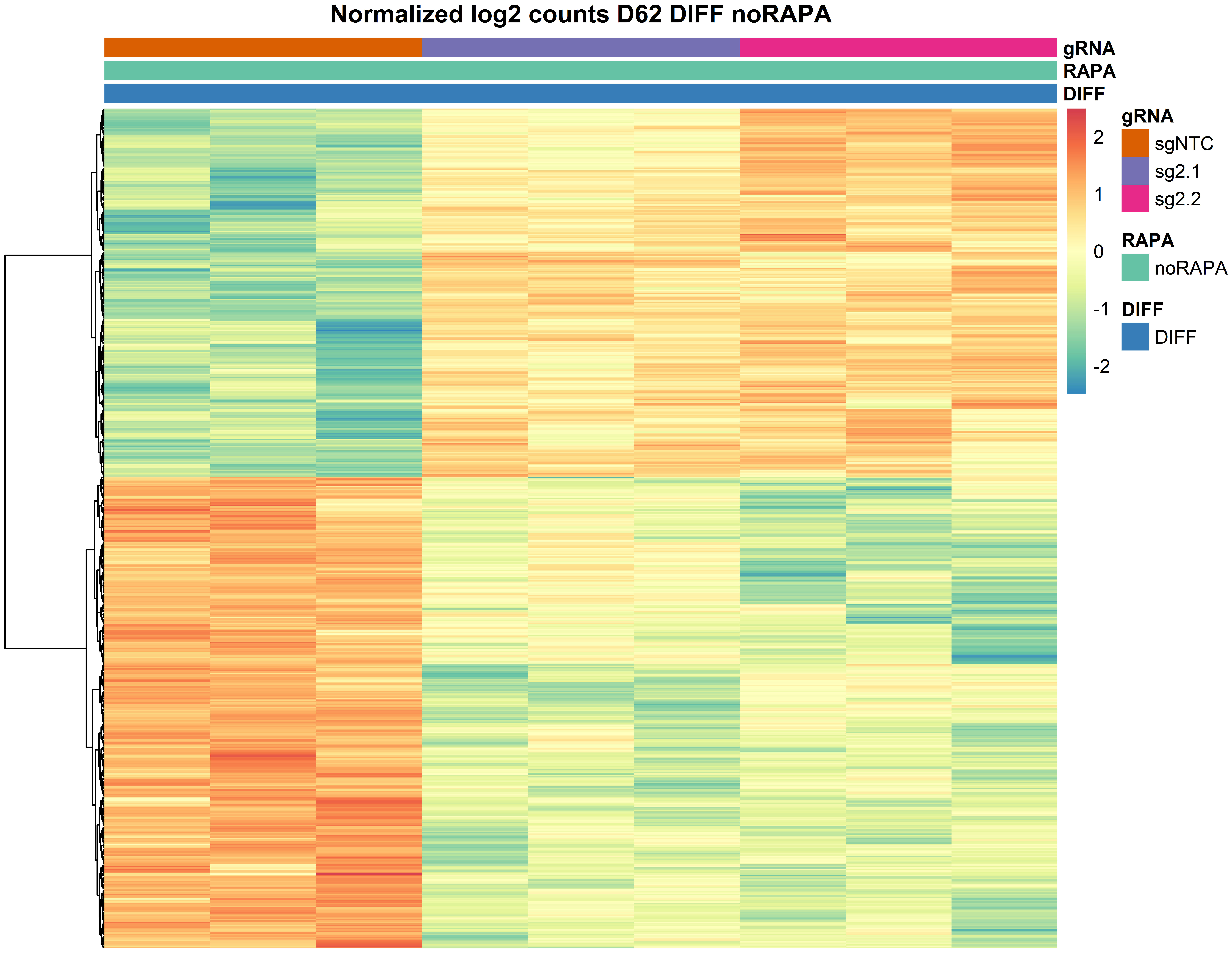
Warning: Removed 3 rows containing missing values (`geom_point()`).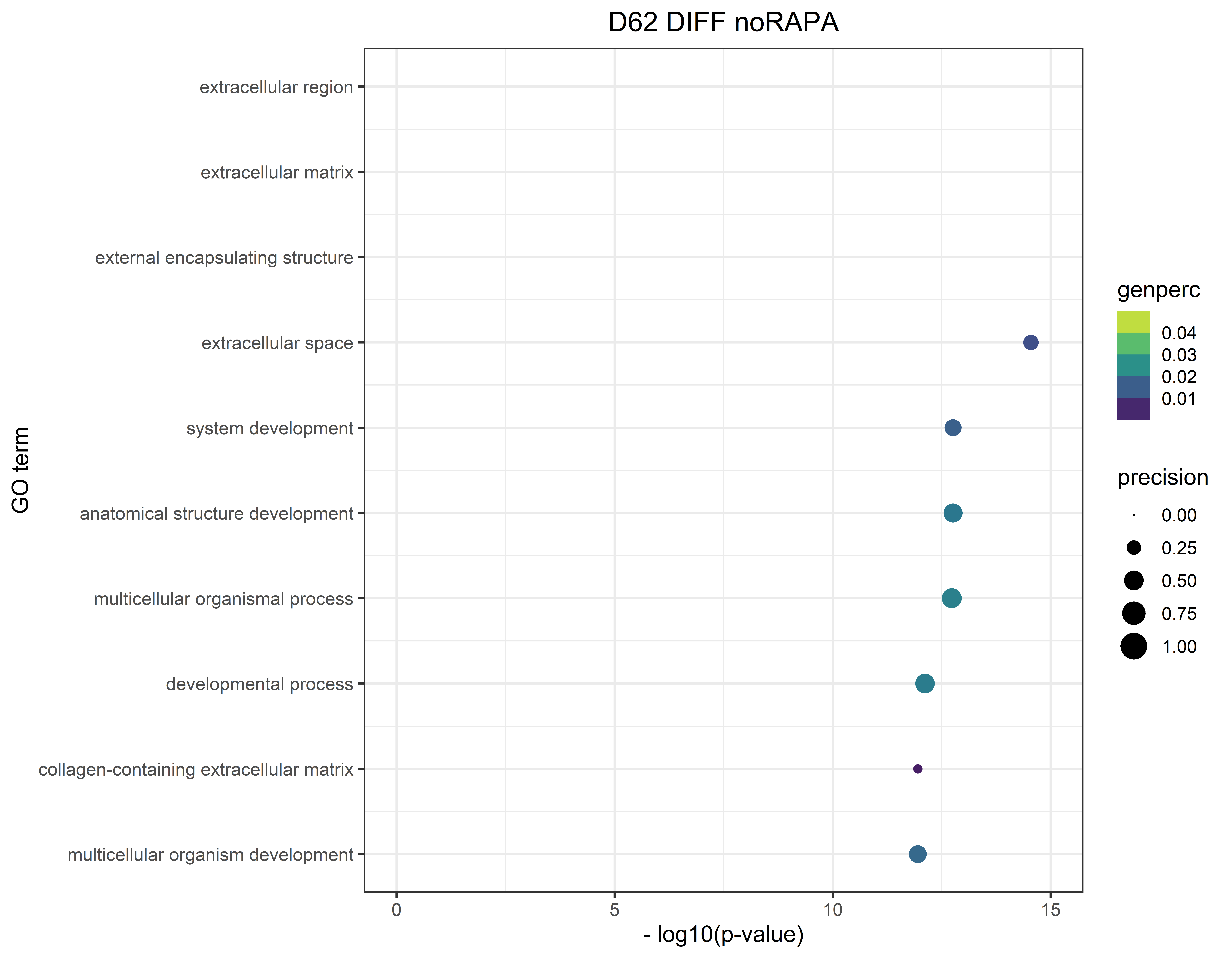
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.6214396 1.18032404 2.187405 2.472160e-03
02_TSC2_KO_Grabole_2016 1.8229487 1.53478736 2.164433 4.166022e-12
03_TSC2_Grabole-2016 1.9023263 1.59233264 2.277780 2.380051e-13
04_TSC2-tuber-Martin2017 3.6773344 1.96824099 6.424719 5.307322e-05
05_TSC2-SEN_SEGA-Martin2017 2.8805306 2.38227452 3.472770 7.223428e-26
06_all_EpilepsyGene 1.0868739 0.62157183 1.781176 6.950876e-01
07_high_conf_EpilepsyGene 1.3861102 0.54274367 2.966400 3.629156e-01
08_ASD_Epilepsy_EpilepsyGene 0.8233931 0.21973239 2.179563 1.000000e+00
09_Epi25_dataset 0.5616319 0.06672542 2.104121 5.862315e-01
10_SFARI_all 1.3179773 0.68673061 2.318193 3.269155e-01
11_SFARI_score4plus 1.3645524 0.63911573 2.590754 3.425416e-01
12_DeRubeis_RDNV 2.2289846 0.86174833 4.859288 4.805024e-02
13_Iossifov_RDNV 1.2967790 0.83783634 1.932040 2.129306e-01
14_FMRP_Darnell 1.4056939 0.99616952 1.939874 4.001085e-02
15_Voineagu_M12 1.0541549 0.57921150 1.779947 7.843163e-01
16_Voineagu_M16 3.3334839 2.31711243 4.692098 7.567759e-10
17_ID_genes_Neel 1.2092138 0.67761522 2.013593 4.780496e-01
18_ID_genes_pinto2014 1.0129237 0.47653675 1.911436 8.702785e-01
19_Fromer_SCZ_FunDN 0.8766391 0.51109705 1.414184 7.286343e-01
20_Cocchi_Pruning 3.9652851 2.00793636 7.242600 8.267264e-05
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.48331438 0.16200274 4.944320e-02
02_TSC2_KO_Grabole_2016 0.60045533 0.08778749 8.332045e-11
03_TSC2_Grabole-2016 0.64307753 0.09075383 4.760102e-12
04_TSC2-tuber-Martin2017 1.30218813 0.31890198 1.061464e-03
05_TSC2-SEN_SEGA-Martin2017 1.05797452 0.09689735 1.444686e-24
06_all_EpilepsyGene 0.08330561 0.28510684 1.000000e+00
07_high_conf_EpilepsyGene 0.32650138 0.47837731 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene -0.19432159 0.67399146 1.000000e+00
09_Epi25_dataset -0.57690860 1.08686773 1.000000e+00
10_SFARI_all 0.27609825 0.33260788 1.000000e+00
11_SFARI_score4plus 0.31082645 0.38698785 1.000000e+00
12_DeRubeis_RDNV 0.80154614 0.48486641 9.610049e-01
13_Iossifov_RDNV 0.25988349 0.22286530 1.000000e+00
14_FMRP_Darnell 0.34053107 0.17569842 8.002171e-01
15_Voineagu_M12 0.05273943 0.30552398 1.000000e+00
16_Voineagu_M16 1.20401797 0.18555929 1.513552e-08
17_ID_genes_Neel 0.18997038 0.29548268 1.000000e+00
18_ID_genes_pinto2014 0.01284086 0.38472005 1.000000e+00
19_Fromer_SCZ_FunDN -0.13165989 0.27527342 1.000000e+00
20_Cocchi_Pruning 1.37757777 0.34717870 1.653453e-03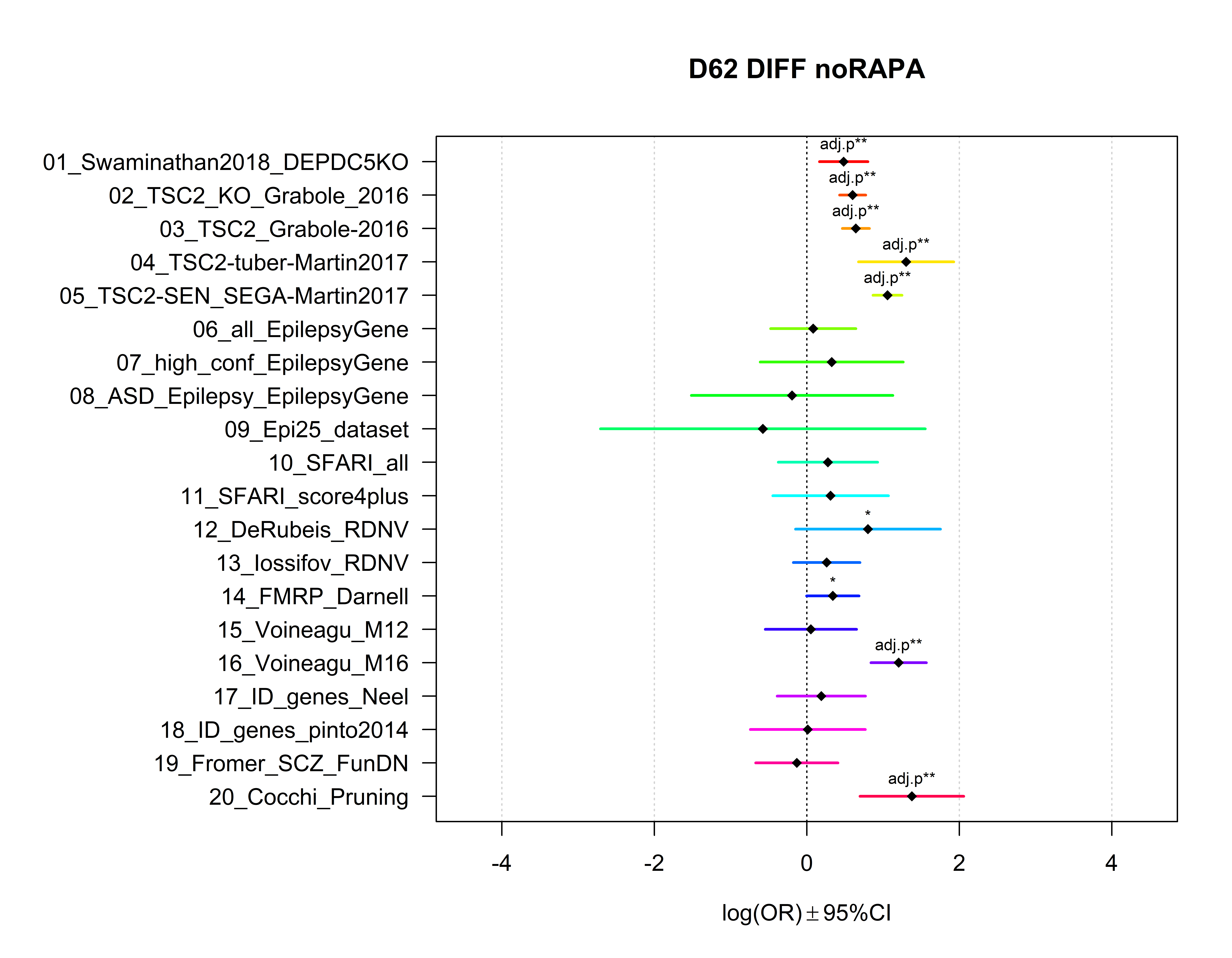
KO effect in noDIFF RAPA
D62
getlinmodoutput(targetdiff = "noDIFF",targetrapa = "RAPA") entrezgene genname log2FC_2.1 fdr_2.1 log2FC_2.2 fdr_2.2
1 375790 AGRN -0.7830596 3.295473e-07 -0.5051920 4.391947e-02
2 2023 ENO1 0.9925868 6.689458e-07 0.6482814 2.479114e-02
3 56998 CTNNBIP1 0.5811856 2.519022e-03 0.5507327 6.635886e-03
4 10630 PDPN 1.4419090 5.919649e-11 0.9738968 4.916388e-05
5 3925 STMN1 -0.6449759 1.811171e-05 -0.6964049 2.409901e-04
6 83667 SESN2 1.4404370 1.129822e-14 1.2728655 6.814788e-08
7 8565 YARS1 1.2112800 1.089305e-04 0.9994600 1.377347e-02
8 4725 NDUFS5 0.8472489 1.037469e-09 0.4949652 3.180663e-02
9 6202 RPS8 -0.2753174 2.813229e-02 -0.4561566 2.444661e-03
10 8503 PIK3R3 1.5760410 6.874351e-20 1.2395143 1.818616e-09
11 9077 DIRAS3 1.7176765 6.602288e-18 1.2839183 2.449181e-07
12 81849 ST6GALNAC5 -1.0478513 1.655478e-13 -0.9760845 1.811898e-09
13 64123 ADGRL4 1.4020313 1.171060e-03 1.2017255 2.361234e-02
14 2633 GBP1 3.9061963 1.681615e-03 4.4053684 3.807240e-05
15 6125 RPL5 -0.6828161 9.147973e-11 -0.4811536 2.981189e-04
16 1301 COL11A1 0.8562904 2.305744e-03 1.1586679 7.574325e-08
17 26227 PHGDH 1.9194937 8.307368e-22 2.3136946 1.106356e-29
18 10628 TXNIP -1.1270322 7.783607e-03 -1.7663456 1.780192e-07
19 140576 S100A16 0.7534756 7.946100e-06 0.5722993 3.517155e-02
20 9181 ARHGEF2 -0.4634304 2.341054e-02 -0.6708935 1.610538e-03
21 10763 NES 0.7072571 3.282436e-08 0.5295565 2.888838e-04
22 9507 ADAMTS4 0.9421749 1.996897e-02 1.1233340 3.324644e-02
23 2212 FCGR2A -2.3711937 5.342599e-05 -2.2818291 6.946215e-04
24 387597 ILDR2 0.4875237 1.697096e-02 0.7833515 2.753216e-05
25 6004 RGS16 1.1311793 8.260652e-10 1.0753088 6.798374e-05
26 116496 NIBAN1 2.0457551 7.102174e-07 1.9144428 7.085667e-05
27 824 CAPN2 0.6197758 1.577312e-02 0.7726694 2.421003e-03
28 149111 CNIH3 1.5732193 1.236103e-07 1.4170177 5.183025e-05
29 3020 H3-3A -0.5390210 4.830611e-03 -0.6040056 5.394219e-03
30 183 AGT 0.8072493 8.005232e-04 0.8280949 2.328836e-04
31 4811 NID1 -1.5016308 2.866951e-03 -1.5060833 1.081566e-02
32 7431 VIM -0.5932418 1.205034e-06 -0.5485910 3.899045e-03
33 8325 FZD8 -0.9417011 1.633203e-02 -1.1975699 1.054004e-02
34 219699 UNC5B 0.8163324 4.047024e-08 0.9264237 1.957529e-08
35 54541 DDIT4 1.9127348 2.163219e-11 1.6434186 1.957529e-08
36 23234 DNAJC9 0.7349550 3.138131e-02 0.8261810 2.794636e-02
37 219654 ZCCHC24 -0.5431326 8.989282e-04 -0.5714200 2.991268e-04
38 9124 PDLIM1 1.2258187 2.605397e-06 1.0595779 1.356536e-03
39 80019 UBTD1 1.2274180 1.396894e-02 1.2299378 3.054954e-02
40 83742 MARVELD1 -1.3485243 3.267090e-06 -0.9735816 8.140662e-03
41 644511 RPL13AP6 -0.7382874 2.389756e-04 -0.5978152 1.973293e-02
42 10581 IFITM2 -1.9856204 8.953844e-17 -2.2578514 8.369471e-18
43 10410 IFITM3 -0.5164410 1.260918e-04 -0.5609883 3.801745e-03
44 977 CD151 0.7671350 2.015220e-06 0.6316686 3.018086e-02
45 8665 EIF3F -0.5480935 1.219357e-03 -0.6635923 1.368846e-03
46 4004 LMO1 -2.5797355 4.047024e-08 -2.6040132 6.128110e-15
47 6157 RPL27A -0.3793310 1.881240e-04 -0.4315367 2.012879e-02
48 10335 IRAG1 1.7877320 4.264072e-03 1.5555086 4.265670e-02
49 5080 PAX6 -0.8436887 1.428381e-02 -0.9330713 1.884973e-02
50 90993 CREB3L1 0.9812654 2.456859e-03 1.3250038 1.482465e-05
51 8525 DGKZ 0.7794046 8.509468e-04 0.7812704 1.054004e-02
52 254263 CNIH2 0.7496305 1.097413e-02 0.8477914 1.874942e-02
53 595 CCND1 2.1436979 2.450562e-06 1.9365598 7.180898e-06
54 23201 FAM168A -0.7612918 3.457406e-03 -0.8598301 4.510339e-04
55 871 SERPINH1 0.4430623 2.882487e-04 0.5165011 6.030246e-03
56 143686 SESN3 1.2348235 2.360075e-07 1.3193109 4.839548e-08
57 57562 CEP126 1.5630160 9.417432e-03 1.4566951 3.855296e-02
58 114908 TMEM123 -0.8543199 2.646062e-03 -0.7028913 4.326217e-02
59 1410 CRYAB 0.7419323 1.195297e-04 0.7986174 3.590581e-05
60 4684 NCAM1 0.5426350 1.034665e-03 0.6015482 6.630190e-04
61 6876 TAGLN -0.5067508 2.953048e-05 -0.4291982 2.361234e-02
62 57453 DSCAML1 1.6431629 3.329927e-02 1.6731081 4.440006e-02
63 50937 CDON -3.7829266 3.157166e-02 -5.2549444 2.627062e-03
64 84623 KIRREL3 -3.7248064 4.168679e-02 -7.5168316 2.404412e-04
65 2113 ETS1 -7.9366484 2.388550e-05 -4.3469543 9.600231e-03
66 27087 B3GAT1 -0.5597962 3.015777e-03 -0.6811093 4.245730e-03
67 894 CCND2 0.8934701 1.992293e-12 1.0170601 2.703638e-11
68 2 A2M -0.6799047 5.819831e-07 -0.8902810 7.029165e-08
69 9976 CLEC2B 4.8120263 1.315740e-04 5.6621290 1.391724e-08
70 2012 EMP1 -1.4949405 1.318950e-07 -1.5188116 3.492679e-06
71 3709 ITPR2 2.0080495 7.922213e-10 2.0133932 2.096133e-08
72 81539 SLC38A1 2.4704960 3.198456e-02 2.8516234 3.339888e-03
73 1280 COL2A1 -1.7108195 1.663978e-10 -1.0952326 3.167592e-06
74 10376 TUBA1B 0.6889250 2.471587e-06 0.4468471 1.089521e-02
75 7846 TUBA1A 0.8326231 9.143257e-16 0.4436578 2.889052e-03
76 84790 TUBA1C 1.1162251 6.938667e-15 1.0612090 5.720949e-07
77 1975 EIF4B -0.5280988 4.341873e-02 -0.8717308 1.239713e-02
78 3178 HNRNPA1 -0.8804545 7.749494e-10 -0.7161885 1.962200e-05
79 84324 SARNP 1.4141260 2.519022e-03 1.7237333 1.345319e-05
80 23344 ESYT1 0.9145527 2.499382e-02 1.2379576 2.421003e-03
81 4666 NACA -0.3619302 2.565329e-02 -0.4303974 2.460459e-02
82 6472 SHMT2 1.1882159 4.748700e-03 1.4569799 4.082249e-04
83 144453 BEST3 1.7109231 1.009329e-09 1.6966933 6.979850e-08
84 4673 NAP1L1 -0.5458068 3.654321e-02 -0.5276290 3.441171e-03
85 160335 TMTC2 -1.1358907 9.907774e-05 -1.4323673 4.235177e-05
86 1848 DUSP6 -0.8163379 3.224719e-10 -0.6578631 4.235177e-05
87 50515 CHST11 -0.4035131 2.813229e-02 -0.4537174 2.227514e-02
88 6311 ATXN2 -0.4682890 3.403559e-02 -0.6283188 1.415230e-02
89 51499 TRIAP1 1.2939512 3.267090e-06 1.1778883 2.091401e-04
90 5027 P2RX7 8.8802685 2.028298e-08 9.2232172 1.275959e-08
91 8848 TSC22D1 0.5955270 3.869239e-05 0.6997709 1.469306e-04
92 9445 ITM2B -0.5793603 3.850154e-06 -0.4741785 1.899613e-03
93 5100 PCDH8 -1.9371643 4.990478e-08 -2.0083000 8.932002e-06
94 8874 ARHGEF7 1.1201255 4.631796e-09 1.1519789 1.864475e-09
95 2621 GAS6 1.9298677 4.443006e-02 2.0729415 4.888677e-02
96 4860 PNP 1.8155705 4.763449e-04 1.3442116 4.867427e-02
97 28526 <NA> 2.1231742 2.861885e-10 2.0565643 4.000953e-09
98 10278 EFS 1.5947969 3.102180e-04 1.4795907 1.384945e-02
99 9985 REC8 1.5232430 8.193040e-20 1.5746823 2.495274e-13
100 4053 LTBP2 -2.8539315 1.681167e-07 -1.2831123 1.524297e-02
101 55384 MEG3 1.0667223 1.742715e-06 1.0011398 2.626418e-04
102 3320 HSP90AA1 0.4304650 1.206121e-04 0.4029921 4.724906e-03
103 1152 CKB 0.9097570 4.479705e-16 0.8155128 4.000953e-09
104 113146 AHNAK2 1.7372416 5.949155e-03 1.5949863 2.606146e-02
105 64397 ZNF106 0.8981486 1.295912e-07 0.7291592 4.765263e-04
106 51065 RPS27L 0.4214371 4.873396e-04 0.4300981 5.846331e-03
107 57611 ISLR2 -1.5551381 3.305725e-04 -1.4950848 6.617584e-04
108 23205 ACSBG1 1.1126580 1.166430e-06 0.9279995 5.090320e-04
109 10509 SEMA4B -0.6743508 8.615007e-03 -0.7813766 1.550838e-02
110 56963 RGMA 1.3836463 7.004728e-18 1.2560281 1.474468e-06
111 7023 TFAP4 -2.1033429 3.075167e-02 -2.5526745 6.030246e-03
112 2013 EMP2 -1.3564345 8.248599e-14 -1.0198586 1.692998e-06
113 124152 IQCK -0.5708228 2.090980e-02 -0.8200022 2.981189e-04
114 26471 NUPR1 2.7862916 8.307368e-22 1.8659235 1.204677e-06
115 11273 ATXN2L -1.2475375 1.486038e-02 -1.1333805 4.149063e-02
116 643911 <NA> 2.5605456 1.390433e-03 2.7846333 4.235177e-05
117 2775 GNAO1 -0.9638596 1.203127e-05 -0.7296164 6.966556e-03
118 23406 COTL1 0.6726662 4.758231e-05 0.5845588 2.415700e-04
119 10381 TUBB3 0.3371715 3.648935e-02 0.4992626 2.098316e-03
120 79007 DBNDD1 1.3741020 1.490219e-09 1.1093693 5.929156e-03
121 482 ATP1B2 -0.4918224 2.015220e-06 -0.5708962 1.147631e-05
122 9423 NTN1 1.2839665 9.804621e-11 0.9844203 2.415700e-04
123 9953 HS3ST3B1 -0.9973668 3.576278e-04 -1.0881248 7.574325e-08
124 256302 NATD1 -1.2628394 1.639927e-03 -1.5612716 4.635308e-03
125 8153 RND2 0.9221021 1.057573e-03 0.7258360 3.527505e-02
126 4830 NME1 1.0416458 5.814657e-07 1.1939895 3.270526e-06
127 9902 MRC2 -0.5129959 1.783518e-02 -0.4305769 1.442417e-02
128 27092 CACNG4 -0.6676901 1.302609e-04 -0.7511143 2.981189e-04
129 23580 CDC42EP4 0.6685190 4.006227e-04 0.5390616 2.108486e-02
130 54549 SDK2 -1.3802060 2.536855e-03 -1.0961501 2.245900e-02
131 51155 JPT1 0.4944537 2.663520e-04 0.6996336 2.207775e-05
132 643008 SMIM5 6.9288262 5.303260e-03 6.9203360 1.168431e-02
133 283991 UBALD2 -0.7119266 5.631216e-05 -0.8115246 1.677474e-05
134 146664 MGAT5B -2.5234477 7.102174e-07 -1.9241422 3.498456e-05
135 9021 SOCS3 -0.9138269 1.947296e-03 -0.9073798 9.070174e-04
136 7077 TIMP2 -0.4551642 1.135516e-04 -0.4129483 8.746301e-03
137 284184 NDUFAF8 0.9130603 7.301495e-07 0.6962730 5.029262e-03
138 71 ACTG1 -0.4620899 5.100470e-05 -0.6467827 6.814788e-08
139 5831 PYCR1 0.9824516 8.417856e-04 0.7612016 4.429016e-02
140 23253 ANKRD12 -0.5791114 1.678079e-02 -0.5240250 3.479436e-02
141 361 AQP4 0.6894458 3.691573e-05 0.7537129 2.238061e-04
142 64762 GAREM1 -2.0889792 4.439245e-05 -1.4588854 3.552128e-03
143 115701 ALPK2 -0.6110394 2.348780e-02 -0.6112116 4.272354e-02
144 23239 PHLPP1 -0.6609033 3.545682e-02 -0.7426337 1.705119e-02
145 92126 DSEL 0.9079642 2.866951e-03 0.9849736 2.213103e-02
146 1265 CNN2 -1.1306687 7.408768e-09 -1.3577834 1.329113e-10
147 1938 EEF2 -0.4387126 1.112270e-02 -0.5091792 8.812568e-03
148 57153 SLC44A2 -0.7954645 2.850143e-06 -1.0322158 9.275215e-07
149 976 ADGRE5 0.9521932 1.500797e-03 1.0512485 5.599683e-03
150 4713 NDUFB7 0.8012245 2.922105e-02 0.9426144 4.888677e-02
151 1463 NCAN 0.4557637 1.755276e-05 0.4400428 2.516062e-02
152 163175 LGI4 -0.8811531 1.229615e-02 -0.9355542 1.937726e-02
153 4793 NFKBIB 1.0886830 1.577312e-02 1.4209632 2.047499e-04
154 115290 FBXO17 4.9836685 1.350014e-03 4.5657529 3.884168e-02
155 10683 DLL3 1.1217341 1.124895e-05 1.2070370 5.387399e-07
156 348 APOE 1.4234066 9.147973e-11 1.3682021 6.814788e-08
157 27113 BBC3 1.3959784 2.022786e-08 1.4153733 1.974888e-03
158 29997 NOP53 -0.4237622 2.877179e-02 -0.5037452 3.082554e-02
159 2014 EMP3 1.6507168 6.222956e-04 1.4941916 8.746301e-03
160 10856 RUVBL2 0.8825740 1.666519e-02 0.9993349 2.076668e-02
161 23040 MYT1L -6.4617983 1.711582e-02 -6.3318910 4.212373e-02
162 129642 MBOAT2 -0.8413429 1.192703e-03 -0.7660251 2.014167e-02
163 28951 TRIB2 -0.5659250 5.319850e-04 -0.3724000 1.526979e-02
164 151354 LRATD1 1.2190968 9.221227e-13 1.2749206 2.495274e-13
165 81553 CYRIA 1.2878219 2.468035e-03 1.4105290 1.540420e-03
166 91461 PKDCC -0.6826956 3.282436e-08 -0.6177082 4.975196e-04
167 23433 RHOQ 0.8040852 8.338466e-04 0.9982660 3.810327e-06
168 618 BCYRN1 2.6693594 2.172692e-38 2.5916460 5.352601e-05
169 53335 BCL11A -1.4688626 8.531540e-05 -1.0771037 1.075762e-02
170 94097 SFXN5 0.9341462 7.350328e-03 0.9365237 7.635315e-03
171 9168 TMSB10 -0.6896943 7.222704e-12 -0.7307418 1.337441e-10
172 56910 STARD7 0.5488665 2.024327e-03 0.4607817 2.664770e-02
173 100506421 PANTR1 -0.7180148 2.850143e-06 -0.6293211 1.532357e-04
174 1622 DBI 0.7281180 4.076437e-12 0.4807958 7.731474e-04
175 6328 SCN3A 1.4516193 7.329401e-11 1.4191266 2.164187e-05
176 6326 SCN2A 2.0080540 6.062901e-03 1.8093617 2.983559e-02
177 165215 FAM171B 0.7655218 1.189111e-03 0.6715021 3.551199e-02
178 3329 HSPD1 0.4468808 7.905957e-03 0.6753641 2.647578e-04
179 26010 SPATS2L 0.6453491 2.024327e-03 0.5706857 2.573422e-02
180 389073 C2orf80 -0.8819781 6.627774e-09 -0.5671427 6.629801e-03
181 7857 SCG2 1.5795399 1.269940e-13 1.2893529 1.552195e-07
182 81618 ITM2C -1.1569375 9.884501e-16 -0.5885298 2.108486e-02
183 2817 GPC1 0.6368793 2.226108e-03 0.6342499 7.826915e-03
184 2280 FKBP1A 0.5414863 8.005232e-04 0.5352480 5.218521e-03
185 5332 PLCB4 -1.9157545 9.856946e-04 -1.3769301 4.239241e-02
186 3397 ID1 3.2466858 5.396866e-05 3.1325201 2.711859e-04
187 4826 NNAT 0.7841971 4.714573e-02 1.4871359 3.871618e-06
188 9334 B4GALT5 0.5299172 2.860703e-02 0.5847839 3.975828e-02
189 3785 KCNQ2 -0.9591275 4.096321e-06 -0.5675444 1.958154e-02
190 1826 DSCAM 2.1433966 4.691324e-08 2.5850132 7.237897e-14
191 6285 S100B -0.3839336 7.350328e-03 -0.4833253 4.816150e-04
192 440823 MIAT 1.2706644 4.198782e-06 0.9692973 7.554949e-03
193 83999 KREMEN1 -1.3918724 1.085639e-03 -1.3988816 2.421003e-03
194 4627 MYH9 -0.5062993 5.165683e-03 -0.5001791 7.528845e-03
195 6122 RPL3 -0.3886845 1.228736e-03 -0.4588551 1.327969e-02
196 10752 CHL1 -0.8898417 2.592465e-10 -0.7759620 5.720949e-07
197 6138 RPL15 -0.7531904 2.929699e-12 -0.3787905 3.493538e-02
198 57060 PCBP4 0.9487725 8.176796e-03 0.9560938 3.160312e-02
199 7086 TKT 0.6878846 3.305725e-04 0.9727439 1.776350e-05
200 54756 IL17RD -0.9355540 3.811236e-12 -0.9427079 4.125136e-10
201 100506621 <NA> 1.6937138 1.091287e-03 2.0728189 4.020557e-06
202 151887 CCDC80 1.1534571 1.116952e-21 0.9686929 6.861728e-07
203 2596 GAP43 1.8580749 2.477698e-66 1.4625612 4.877498e-32
204 4171 MCM2 0.8059128 2.288285e-03 0.7859034 2.344828e-02
205 2047 EPHB1 2.7484413 4.873700e-08 2.9778188 7.221653e-09
206 5947 RBP1 -0.4957535 1.305523e-02 -0.6533588 3.243665e-03
207 64393 ZMAT3 0.7323454 1.007526e-05 0.8285674 8.066086e-06
208 2049 EPHB3 -0.8126826 3.015777e-03 -1.0063983 2.781484e-05
209 3280 HES1 -0.5886471 2.414069e-04 -0.5465658 9.932744e-03
210 347 APOD -3.0953962 4.034502e-07 -2.2712642 2.177719e-02
211 6165 RPL35A -0.3580130 5.165683e-03 -0.4266551 1.974888e-03
212 57537 SORCS2 -0.6450856 2.461379e-02 -0.8989371 3.458917e-03
213 23231 SEL1L3 -0.5207816 2.813229e-02 -0.5739710 2.014167e-02
214 5156 PDGFRA -0.9392241 1.186302e-09 -0.9665092 4.020557e-06
215 3490 IGFBP7 1.1534380 2.344637e-12 1.2542994 4.782721e-12
216 55752 SEPTIN11 0.4851816 1.063820e-02 0.4679387 4.017480e-02
217 79966 SCD5 0.3272269 1.844456e-02 0.4102574 1.906731e-02
218 8404 SPARCL1 2.8087175 1.905333e-14 1.8385528 8.626136e-07
219 6164 RPL34 -0.4329017 1.601551e-04 -0.6035140 4.510339e-04
220 4889 NPY5R -1.5087702 8.027025e-03 -1.6376177 8.603475e-03
221 55714 TENM3 -2.2138623 1.449066e-09 -1.0767567 5.441991e-03
222 10409 BASP1 -0.8340552 2.090980e-02 -0.8739130 7.329230e-03
223 6897 TARS1 0.7890136 2.016526e-03 0.8043556 1.402495e-04
224 689 BTF3 -0.8444687 1.855508e-06 -0.4459926 4.748658e-02
225 22836 RHOBTB3 -0.4838039 4.769316e-04 -0.4630598 8.814686e-03
226 401207 C5orf63 1.6985122 3.646139e-14 1.8987112 1.026370e-17
227 84466 MEGF10 -1.4201656 1.901231e-07 -1.5194971 4.775096e-09
228 9547 CXCL14 1.6189268 2.189710e-09 1.6657979 9.439242e-09
229 56662 VTRNA1-3 6.8055300 1.097430e-02 7.2568102 1.959645e-03
230 81848 SPRY4 -1.5145855 4.034502e-07 -1.1529884 4.816150e-04
231 202374 STK32A 1.6836929 2.536855e-03 2.1266387 1.002760e-06
232 85027 SMIM3 1.1232749 3.806986e-05 0.9346778 1.000157e-03
233 10569 SLU7 -0.6878420 1.590930e-02 -0.7179102 5.181694e-03
234 3779 KCNMB1 -1.6471229 2.618520e-02 -2.7077959 3.017350e-03
235 100113407 TMEM170B -1.4196314 7.141358e-04 -1.2076728 1.054004e-02
236 7726 TRIM26 5.8706542 2.415968e-02 6.3924411 6.381110e-03
237 1026 CDKN1A 1.3714055 1.562515e-37 1.1333855 5.810533e-19
238 1915 EEF1A1 -0.6087627 2.446383e-10 -0.7138034 2.001388e-04
239 5570 PKIB 1.3849946 7.563702e-03 1.8809301 1.404530e-07
240 1490 CCN2 -3.0518736 3.059983e-15 -2.9008147 2.393523e-15
241 6624 FSCN1 -0.3265981 1.854946e-02 -0.4506110 8.613338e-03
242 221833 SP8 -1.0011656 2.223689e-06 -0.7832690 2.091401e-04
243 4852 NPY -1.6483751 1.819183e-06 -1.3018714 1.352032e-04
244 3181 HNRNPA2B1 -0.4005258 3.193098e-02 -0.5780810 1.368846e-03
245 9586 CREB5 0.8936772 9.713426e-03 0.7844193 3.565763e-02
246 9865 TRIL -0.9675944 1.594456e-03 -0.7852621 4.176142e-02
247 107 ADCY1 -1.2156447 2.932513e-03 -1.1682966 2.877580e-02
248 2970 <NA> 0.5622302 5.236522e-04 0.5116459 8.635876e-03
249 2791 GNG11 0.7480073 3.497644e-03 0.9998186 2.546048e-03
250 23089 PEG10 0.8827558 4.795695e-10 0.9037230 7.574325e-08
251 1749 DLX5 -2.1536790 1.706705e-05 -1.7130560 3.436082e-03
252 25798 BRI3 -0.5524484 1.082603e-05 -0.6486295 8.919168e-06
253 11333 PDAP1 0.6446609 1.485309e-03 0.5063778 3.975828e-02
254 5054 SERPINE1 -1.5376214 1.512854e-09 -1.3573869 8.325880e-08
255 7425 VGF 0.9465103 6.958135e-03 0.8113525 2.117367e-02
256 4232 MEST -0.6744520 7.800528e-03 -0.8458414 2.354501e-03
257 26958 COPG2 -2.2304260 2.247738e-04 -1.6123694 2.402438e-02
258 5420 PODXL -0.5721857 2.519022e-03 -0.6268907 4.708311e-03
259 5799 PTPRN2 -0.9268557 9.985470e-08 -0.9405245 2.514684e-04
260 4023 LPL -1.2582515 7.773822e-04 -1.8384670 3.834286e-05
261 4741 NEFM 3.0095868 3.973061e-18 2.2470007 9.724940e-07
262 4747 NEFL 1.5865818 3.491512e-15 1.6747982 3.710007e-14
263 665 BNIP3L -0.7188864 6.475075e-03 -0.7062358 5.350728e-03
264 81551 STMN4 2.6841855 6.874351e-20 2.7015071 5.810533e-19
265 2185 PTK2B 1.6216028 3.574477e-11 1.3509460 4.955018e-06
266 55893 ZNF395 -1.0583604 3.545108e-04 -0.9710353 4.202729e-03
267 5327 PLAT 0.5622500 9.144476e-04 0.5477463 4.738409e-03
268 6224 RPS20 -0.2514251 4.898070e-02 -0.4470762 2.939269e-03
269 6386 SDCBP -0.6610444 3.873406e-05 -0.5823109 4.753488e-04
270 286183 NKAIN3 0.6304030 1.678079e-02 0.6069123 4.239241e-02
271 79776 ZFHX4 -0.8580157 1.904816e-04 -0.9754281 9.135750e-03
272 11075 STMN2 -5.4518653 9.184294e-03 -7.1246314 1.854458e-03
273 5375 PMP2 -0.8541614 7.408768e-09 -0.5263599 3.662117e-02
274 1666 DECR1 0.6149645 2.161658e-03 0.6789573 2.728435e-02
275 6156 RPL30 -0.4304568 4.502108e-04 -0.5118099 3.229880e-05
276 26986 PABPC1 -0.9318235 6.666927e-23 -0.4246663 2.869663e-03
277 3646 EIF3E -0.8996928 3.831524e-05 -0.5621298 7.433963e-03
278 55638 SYBU 2.5971840 7.922213e-10 1.6174539 2.070713e-02
279 123 PLIN2 0.4829531 1.082883e-02 0.5295214 2.218859e-02
280 6194 RPS6 -0.5083678 4.229489e-06 -0.3805450 3.140998e-02
281 152007 GLIPR2 0.9565742 2.109357e-05 0.8838724 9.420653e-04
282 9568 GABBR2 1.8796693 7.588301e-22 1.7156601 2.495274e-13
283 114299 <NA> 2.7099434 2.122430e-02 2.4605050 2.067453e-02
284 114991 ZNF618 -1.5609130 2.353112e-07 -1.0419385 1.052067e-03
285 85301 COL27A1 -0.9741186 4.873700e-08 -0.7987359 7.586876e-04
286 3371 TNC 1.1052895 8.094478e-08 0.8112919 1.891807e-03
287 23245 ASTN2 2.1917557 1.305523e-02 2.4061737 7.202853e-03
288 1620 BRINP1 -5.6782116 4.292098e-07 -2.2762204 3.934795e-02
289 445 ASS1 1.0118493 1.294548e-06 0.6957962 1.369574e-02
290 10439 OLFM1 -1.1428477 2.632653e-02 -2.1790603 2.938566e-03
291 4571 <NA> 0.8904569 4.633292e-04 0.9738771 1.204677e-06
292 6303 SAT1 -0.6831722 6.662148e-06 -0.6646103 2.041303e-03
293 8406 SRPX 1.1222889 2.424502e-02 1.1012305 3.082554e-02
294 3476 IGBP1 -0.9228563 2.536855e-03 -0.8690712 1.290846e-03
295 6191 RPS4X -0.5594572 1.605964e-07 -0.3573592 3.471283e-02
296 56062 KLHL4 1.1732907 2.683423e-03 1.4676162 4.571160e-06
297 56849 TCEAL7 1.3536698 1.953442e-06 1.4710967 3.728234e-07
298 51186 TCEAL9 1.0009871 2.684026e-20 0.8557534 4.827557e-07
299 5631 PRPS1 1.3766310 2.068477e-05 1.1167823 6.030246e-03
300 10178 TENM1 -1.0166588 1.594456e-03 -1.0612770 3.171332e-02
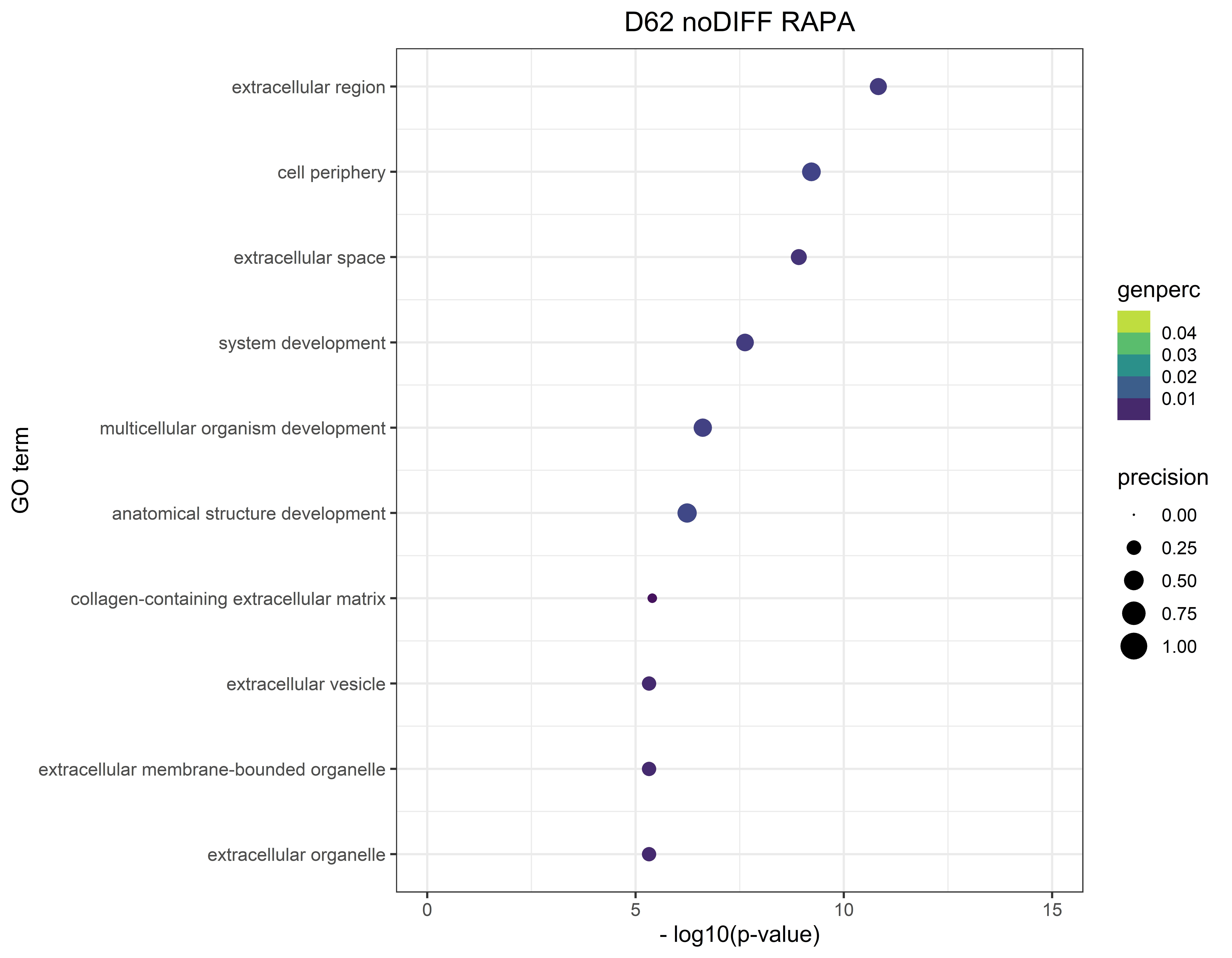
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.4122371 0.88398103 2.161751 1.064169e-01
02_TSC2_KO_Grabole_2016 2.6174092 2.06395627 3.326171 3.787141e-16
03_TSC2_Grabole-2016 2.6248072 2.02776449 3.423101 4.872073e-15
04_TSC2-tuber-Martin2017 3.5444208 1.47664996 7.341278 2.986753e-03
05_TSC2-SEN_SEGA-Martin2017 2.8103010 2.16714292 3.622678 3.947596e-14
06_all_EpilepsyGene 1.3510605 0.66107574 2.482583 2.876916e-01
07_high_conf_EpilepsyGene 1.4989027 0.39925567 3.979140 3.507142e-01
08_ASD_Epilepsy_EpilepsyGene 1.1904707 0.24050357 3.602881 7.426548e-01
09_Epi25_dataset 0.5378743 0.01341566 3.103601 1.000000e+00
10_SFARI_all 0.9435781 0.30133921 2.258520 1.000000e+00
11_SFARI_score4plus 1.0177388 0.27243686 2.680180 7.995110e-01
12_DeRubeis_RDNV 1.7517066 0.35150628 5.363974 2.544902e-01
13_Iossifov_RDNV 1.2685810 0.67973209 2.184355 3.558253e-01
14_FMRP_Darnell 1.8457920 1.20408971 2.735010 4.597121e-03
15_Voineagu_M12 0.6545800 0.20957896 1.560245 4.545189e-01
16_Voineagu_M16 4.1441637 2.63976567 6.279853 7.084273e-09
17_ID_genes_Neel 1.1407571 0.48358037 2.305845 6.974887e-01
18_ID_genes_pinto2014 1.1610513 0.41829347 2.596473 6.521925e-01
19_Fromer_SCZ_FunDN 1.6455723 0.93712403 2.710946 5.712343e-02
20_Cocchi_Pruning 2.6608085 0.83625535 6.527269 4.713633e-02
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.34517503 0.2390279 1.000000e+00
02_TSC2_KO_Grabole_2016 0.96218498 0.1212042 7.574283e-15
03_TSC2_Grabole-2016 0.96500745 0.1316702 9.744146e-14
04_TSC2-tuber-Martin2017 1.26537475 0.4467341 5.973507e-02
05_TSC2-SEN_SEGA-Martin2017 1.03329161 0.1325928 7.895192e-13
06_all_EpilepsyGene 0.30088983 0.3646820 1.000000e+00
07_high_conf_EpilepsyGene 0.40473328 0.6749421 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene 0.17434881 0.8160047 1.000000e+00
09_Epi25_dataset -0.62013038 1.8832665 1.000000e+00
10_SFARI_all -0.05807614 0.5823687 1.000000e+00
11_SFARI_score4plus 0.01758329 0.6724141 1.000000e+00
12_DeRubeis_RDNV 0.56059052 0.8194481 1.000000e+00
13_Iossifov_RDNV 0.23789893 0.3183446 1.000000e+00
14_FMRP_Darnell 0.61290845 0.2179513 9.194242e-02
15_Voineagu_M12 -0.42376150 0.5810680 1.000000e+00
16_Voineagu_M16 1.42170100 0.2301076 1.416855e-07
17_ID_genes_Neel 0.13169214 0.4378724 1.000000e+00
18_ID_genes_pinto2014 0.14932586 0.5208663 1.000000e+00
19_Fromer_SCZ_FunDN 0.49808824 0.2872591 1.000000e+00
20_Cocchi_Pruning 0.97863001 0.5905364 9.427265e-01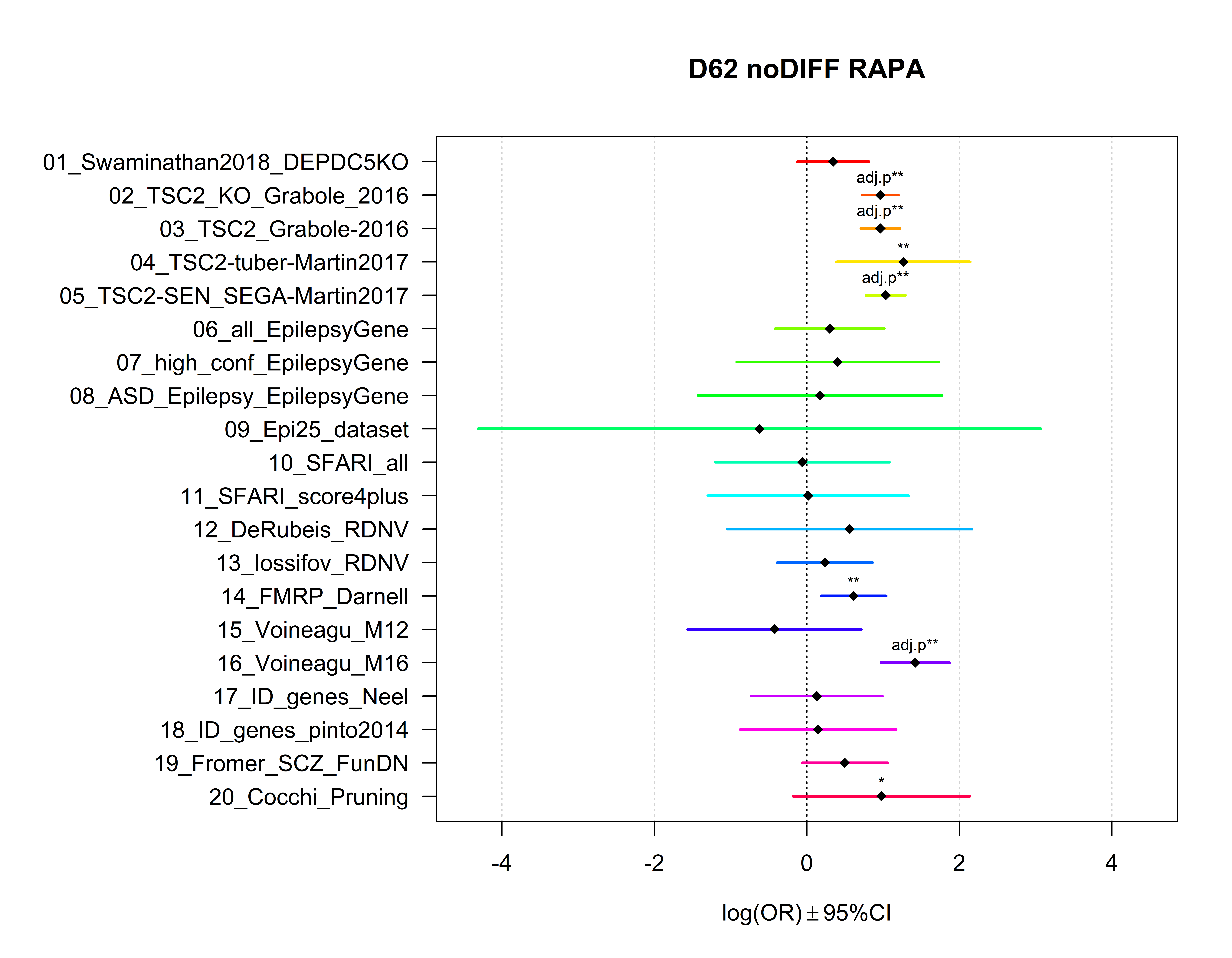
getlinmodoutput(targetdiff = "noDIFF",targetrapa = "RAPA", refset = "NTCwRAPA" ) entrezgene genname log2FC_2.1 fdr_2.1 log2FC_2.2 fdr_2.2
1 64856 VWA1 0.8030245 1.460522e-02 0.8573359 2.379033e-02
2 3925 STMN1 -0.5621997 5.931549e-03 -0.6136263 1.933759e-02
3 9077 DIRAS3 1.9834594 3.993096e-17 1.5497197 2.141006e-07
4 64123 ADGRL4 1.8591906 7.536418e-05 1.6588420 3.756279e-03
5 6004 RGS16 1.1260514 1.116405e-06 1.0703477 2.320961e-03
6 149111 CNIH3 1.5152862 4.321969e-05 1.3591838 3.712089e-03
7 642938 INSYN2A 0.8676982 7.698076e-03 1.0093673 3.529990e-03
8 10581 IFITM2 -1.3183795 7.206153e-04 -1.5901730 1.290538e-04
9 4004 LMO1 -2.2592955 1.057398e-04 -2.2842552 1.029016e-08
10 960 CD44 1.4783297 1.242470e-07 1.3181342 5.315257e-05
11 1410 CRYAB 0.6528849 7.686252e-03 0.7095910 3.377235e-03
12 3709 ITPR2 1.4736906 7.945057e-05 1.4792115 6.769939e-04
13 84790 TUBA1C 0.8353178 1.261909e-05 0.7806232 1.222516e-02
14 160335 TMTC2 -0.9795429 8.552302e-03 -1.2761142 4.320227e-03
15 105376684 <NA> -7.3611655 7.343363e-03 -7.2311260 1.744505e-02
16 28526 <NA> 1.8017386 3.332743e-05 1.7352076 2.424211e-04
17 771 CA12 -0.8196275 2.710179e-04 -0.7915473 2.688799e-03
18 57611 ISLR2 -1.4079246 2.358940e-02 -1.3474653 4.246075e-02
19 56963 RGMA 1.1731544 1.258364e-08 1.0455090 3.529990e-03
20 2013 EMP2 -1.0859712 1.194183e-05 -0.7493947 3.566127e-02
21 643911 <NA> 2.7470949 2.482146e-02 2.9738926 4.672450e-03
22 2775 GNAO1 -1.1535874 8.715183e-06 -0.9192826 4.124693e-03
23 55273 TMEM100 6.4811644 1.942508e-06 5.8792099 9.207100e-04
24 11031 RAB31 0.4843884 1.403435e-02 0.5239510 1.724623e-02
25 361 AQP4 0.7596383 2.396159e-03 0.8238998 4.672450e-03
26 1995 ELAVL3 -0.7733073 5.793295e-03 -0.7804627 2.724563e-02
27 348 APOE 1.1037031 5.920378e-05 1.0484969 2.320961e-03
28 79784 MYH14 -3.8513216 3.308529e-07 -2.3025907 2.636260e-02
29 151354 LRATD1 0.9477041 2.124582e-05 1.0035972 1.153129e-05
30 10267 RAMP1 1.2201730 9.879553e-07 1.0255545 3.256744e-04
31 1827 RCAN1 0.6685747 9.436088e-03 0.7360705 8.953265e-03
32 1826 DSCAM 1.3460266 7.989576e-03 1.7874228 6.724688e-06
33 2596 GAP43 1.3243424 2.481808e-23 0.9288546 1.029016e-08
34 5947 RBP1 -0.6398179 3.926367e-02 -0.7974155 1.173735e-02
35 440982 <NA> 7.2398808 5.793295e-03 7.5740840 2.320961e-03
36 4071 TM4SF1 1.8426096 6.993979e-07 1.4587178 3.712089e-03
37 3280 HES1 -0.7476753 8.866699e-04 -0.7054720 1.438330e-02
38 3490 IGFBP7 2.0336088 4.414861e-30 2.1344478 4.787056e-29
39 79966 SCD5 0.5729957 1.328968e-03 0.6559705 2.080349e-03
40 8404 SPARCL1 2.6761967 3.326139e-08 1.7055759 5.605707e-03
41 401207 C5orf63 0.6795351 2.813485e-02 0.8797647 1.201939e-03
42 85027 SMIM3 0.9774337 2.531243e-03 0.7887911 2.483687e-02
43 1026 CDKN1A 1.0856110 8.078057e-15 0.8475918 1.023411e-06
44 4188 MDFI -0.9871920 4.692325e-02 -1.1913176 2.815769e-02
45 4852 NPY -1.9180616 9.454048e-08 -1.5715347 5.042983e-06
46 23089 PEG10 0.7155489 1.600641e-03 0.7365073 6.201484e-03
47 1749 DLX5 -2.2855033 8.297120e-05 -1.8452078 9.203312e-03
48 25798 BRI3 -0.4903052 8.346773e-03 -0.5864822 4.672450e-03
49 5054 SERPINE1 1.9304914 3.332743e-05 2.1117923 1.740549e-06
50 4747 NEFL 1.2018058 1.301402e-05 1.2900633 1.519552e-05
51 2185 PTK2B 1.6732904 4.938218e-11 1.4023571 1.153129e-05
52 5327 PLAT 0.5947832 5.991585e-03 0.5803625 1.933759e-02
53 11075 STMN2 -6.0356503 7.560988e-03 -7.7099259 1.526840e-03
54 2171 FABP5 1.5497408 2.529428e-06 1.0759024 4.506761e-02
55 9568 GABBR2 1.7738821 1.205907e-16 1.6094149 1.711089e-09
56 56548 CHST7 1.0834089 4.020573e-04 1.1287904 2.320961e-03
57 5358 PLS3 1.0402234 5.747932e-04 1.3025474 1.023411e-06
58 10178 TENM1 -1.3297755 1.057398e-04 -1.3748241 7.463147e-03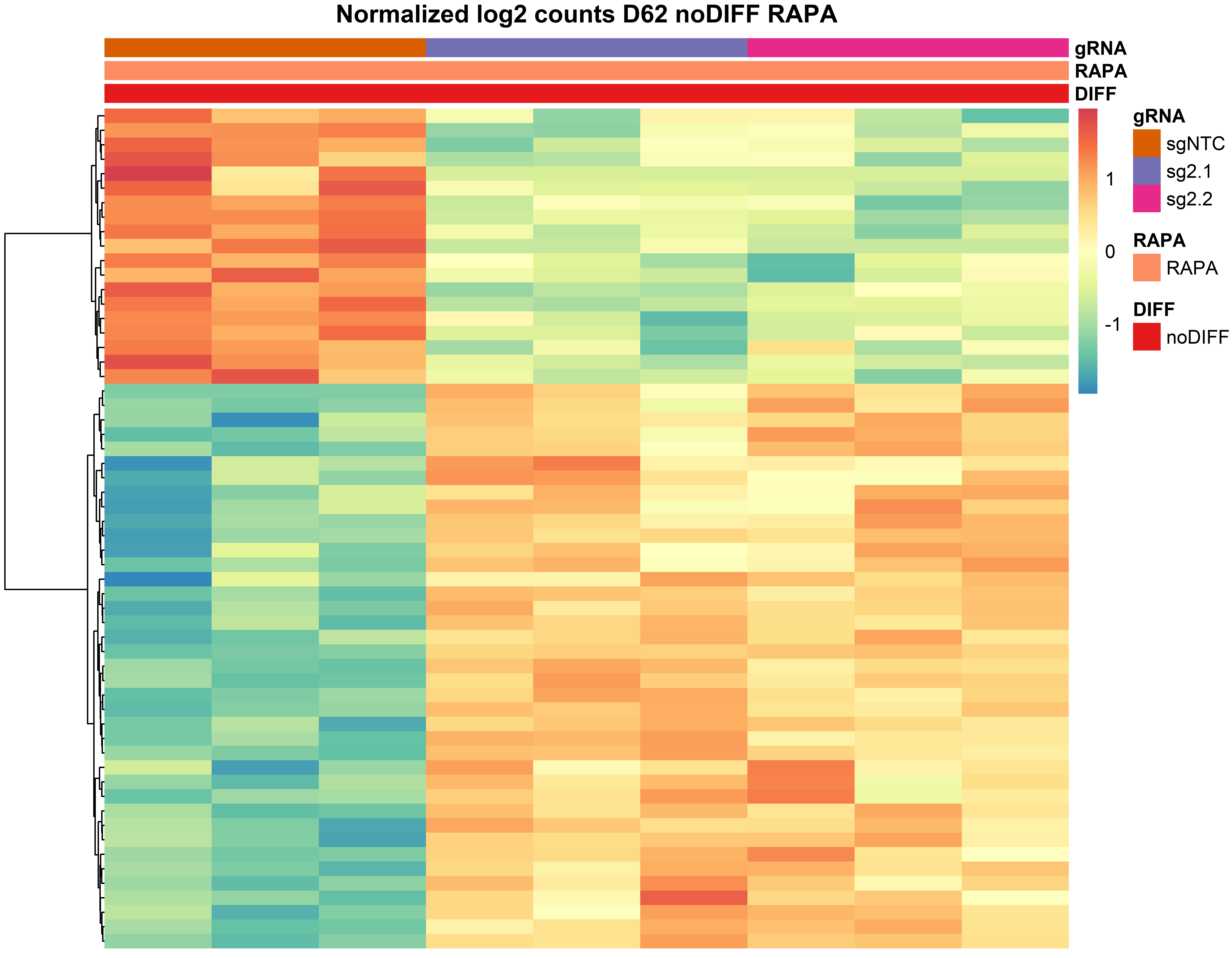
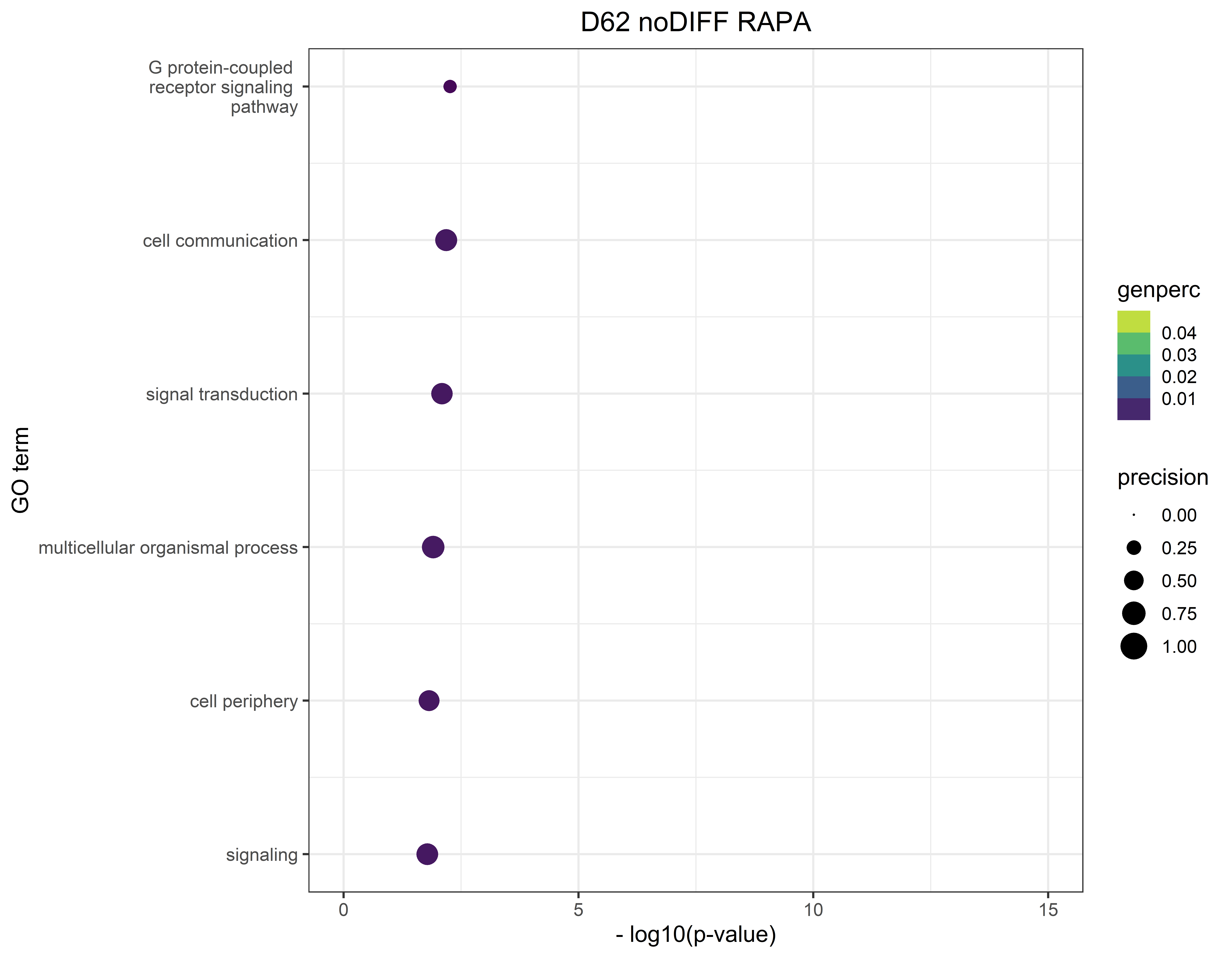
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 0.8777097 0.17523600 2.712007 1.000000e+00
02_TSC2_KO_Grabole_2016 2.3526365 1.35658314 4.115526 1.290207e-03
03_TSC2_Grabole-2016 2.2684120 1.26932030 4.208189 3.500225e-03
04_TSC2-tuber-Martin2017 6.8474658 1.34970593 21.614783 1.149345e-02
05_TSC2-SEN_SEGA-Martin2017 3.6778309 2.05633663 6.438719 7.633899e-06
06_all_EpilepsyGene 1.9288523 0.38423012 5.982515 2.151638e-01
07_high_conf_EpilepsyGene 1.9325519 0.04771494 11.405736 4.097788e-01
08_ASD_Epilepsy_EpilepsyGene 4.2468457 0.49608257 16.421962 8.619854e-02
09_Epi25_dataset 0.0000000 0.00000000 10.853125 1.000000e+00
10_SFARI_all 3.0654629 0.60914585 9.546760 8.239215e-02
11_SFARI_score4plus 2.7072417 0.31756827 10.389322 1.762594e-01
12_DeRubeis_RDNV 0.0000000 0.00000000 11.411849 1.000000e+00
13_Iossifov_RDNV 0.9199100 0.10843513 3.498929 1.000000e+00
14_FMRP_Darnell 1.9630980 0.68678990 4.589240 1.383517e-01
15_Voineagu_M12 0.0000000 0.00000000 2.557987 4.062768e-01
16_Voineagu_M16 4.6064570 1.60576168 10.826397 3.020582e-03
17_ID_genes_Neel 0.0000000 0.00000000 2.729480 6.482452e-01
18_ID_genes_pinto2014 0.0000000 0.00000000 3.735162 6.281407e-01
19_Fromer_SCZ_FunDN 2.0097658 0.52645246 5.482875 1.520947e-01
20_Cocchi_Pruning 2.6773201 0.06590805 15.920384 3.176479e-01
Beta SE Padj
01_Swaminathan2018_DEPDC5KO -0.13043934 0.8220318 1.000000000
02_TSC2_KO_Grabole_2016 0.85553663 0.2809018 0.025804142
03_TSC2_Grabole-2016 0.81908004 0.2962237 0.070004505
04_TSC2-tuber-Martin2017 1.92387863 0.8285673 0.229869090
05_TSC2-SEN_SEGA-Martin2017 1.30232316 0.2966312 0.000152678
06_all_EpilepsyGene 0.65692514 0.8231830 1.000000000
07_high_conf_EpilepsyGene 0.65884134 1.8884449 1.000000000
08_ASD_Epilepsy_EpilepsyGene 1.44617652 1.0955048 1.000000000
09_Epi25_dataset -Inf NaN 1.000000000
10_SFARI_all 1.12019860 0.8244368 1.000000000
11_SFARI_score4plus 0.99593029 1.0933636 1.000000000
12_DeRubeis_RDNV -Inf NaN 1.000000000
13_Iossifov_RDNV -0.08347946 1.0908795 1.000000000
14_FMRP_Darnell 0.67452385 0.5358422 1.000000000
15_Voineagu_M12 -Inf NaN 1.000000000
16_Voineagu_M16 1.52745901 0.5376841 0.060411632
17_ID_genes_Neel -Inf NaN 1.000000000
18_ID_genes_pinto2014 -Inf NaN 1.000000000
19_Fromer_SCZ_FunDN 0.69801819 0.6834757 1.000000000
20_Cocchi_Pruning 0.98481635 1.8899547 1.000000000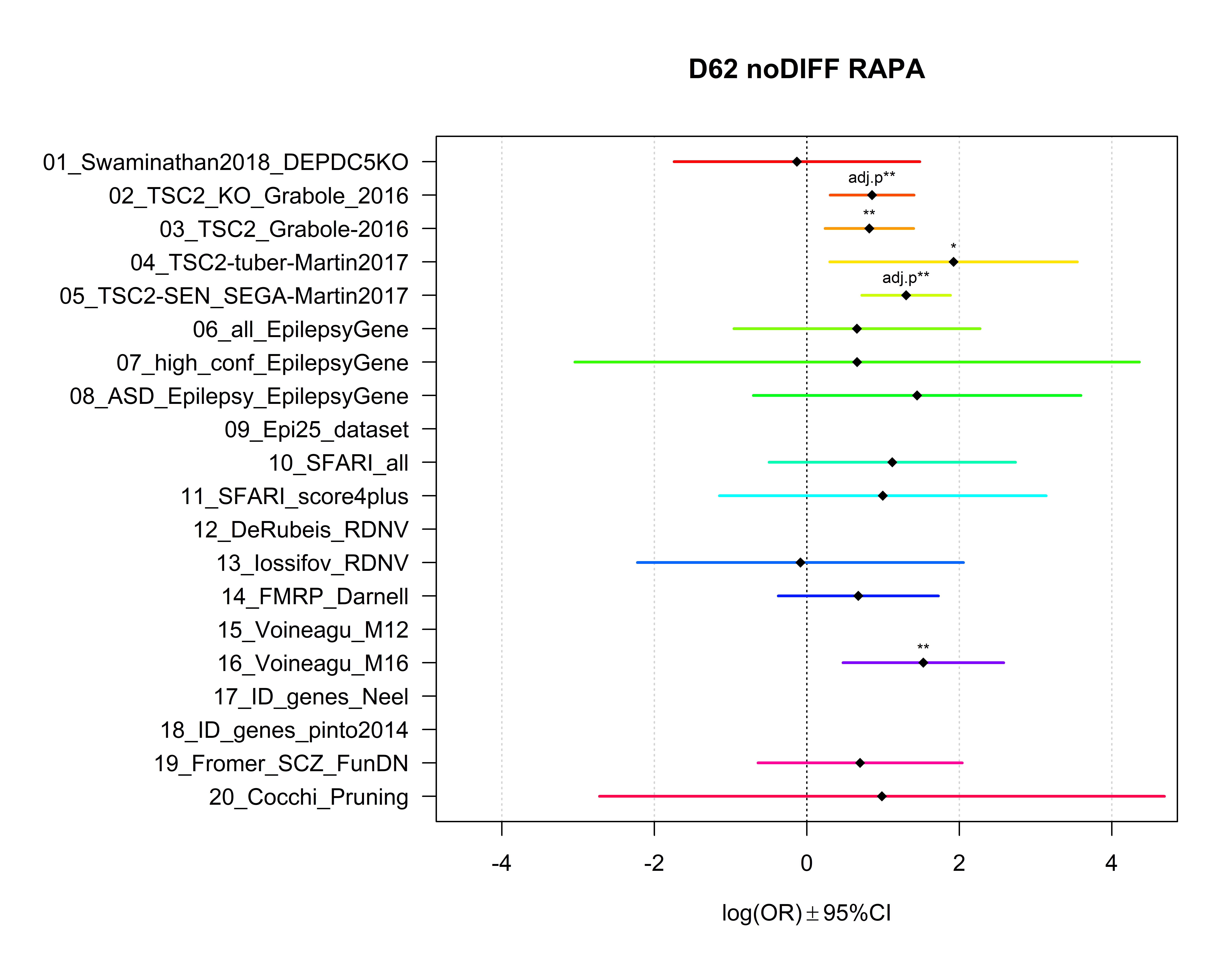
KO effect in DIFF RAPA
D62
getlinmodoutput(targetdiff = "DIFF",targetrapa = "RAPA") entrezgene genname log2FC_2.1 fdr_2.1 log2FC_2.2 fdr_2.2
1 9249 DHRS3 -0.8702049 9.794695e-06 -1.0401296 1.908794e-04
2 10630 PDPN -0.6420981 1.328380e-03 -0.9166837 2.782742e-04
3 126917 IFFO2 -0.7095883 1.328380e-03 -1.0998573 1.808327e-03
4 400745 SH2D5 6.6421948 1.636217e-02 7.6078463 2.379146e-04
5 1889 ECE1 -1.2879425 5.448953e-03 -1.3959975 3.707763e-03
6 2048 EPHB2 -0.8119037 2.074310e-06 -1.1428148 5.146040e-07
7 57134 MAN1C1 -0.6174277 2.778678e-02 -1.1726702 2.003632e-05
8 57190 SELENON -0.8174618 1.048372e-04 -1.0854637 5.790464e-05
9 27245 AHDC1 -0.7011670 4.789158e-02 -1.1058652 3.967818e-03
10 2537 IFI6 -0.9154056 1.733711e-07 -0.9958707 6.209363e-05
11 83667 SESN2 -0.8698284 3.348300e-02 -1.2970142 1.683733e-04
12 1912 PHC2 -0.6629031 2.761389e-02 -0.7101349 3.108137e-02
13 127687 C1orf122 -0.5512172 2.309092e-02 -0.6033709 3.016985e-02
14 84879 MFSD2A 1.5280304 1.535221e-09 2.0551098 6.811424e-17
15 6513 SLC2A1 -0.7119057 2.345858e-02 -0.8277294 2.213270e-02
16 8503 PIK3R3 -0.7119493 4.571128e-02 -1.1458757 3.639720e-03
17 5865 RAB3B 4.5771671 3.571622e-10 5.4144750 1.259123e-04
18 8613 PLPP3 0.5750940 1.442192e-02 0.9404329 1.534580e-04
19 4774 NFIA 0.6128494 2.302822e-03 0.7970079 2.215872e-03
20 79971 WLS 0.9158724 1.021470e-09 1.3217411 8.554430e-11
21 3491 CCN1 1.7166363 1.488136e-23 1.5766023 1.358691e-10
22 23285 <NA> 4.5060199 3.309995e-02 5.0647370 1.606855e-02
23 54874 FNBP1L 0.9193797 4.642875e-02 1.5231530 1.904779e-03
24 9890 PLPPR4 -3.1592433 2.260743e-04 -2.5216635 3.575649e-03
25 127003 CFAP276 1.2241631 5.957958e-03 1.9954874 6.633816e-06
26 1117 CHI3L2 -2.3928183 2.130579e-06 -2.5396234 9.319774e-08
27 7482 WNT2B -1.7703546 4.673293e-03 -2.7979231 9.683555e-07
28 54879 ST7L -1.2524136 3.332666e-03 -1.4958733 6.932722e-03
29 389 RHOC -0.6795066 1.094448e-02 -0.8842241 1.047794e-02
30 476 ATP1A1 -0.8723541 9.694798e-04 -0.8590712 2.055163e-02
31 5738 PTGFRN -0.8139259 4.799805e-03 -1.1677343 8.167610e-05
32 647135 SRGAP2B 1.0715478 6.256772e-03 1.6685860 4.777650e-06
33 6282 S100A11 -0.9719507 3.177550e-06 -1.5945948 7.484662e-11
34 140576 S100A16 -1.0554327 6.375028e-05 -0.9969635 2.319788e-03
35 6284 S100A13 -0.9804621 2.060921e-03 -1.1777575 2.697710e-04
36 63827 BCAN -1.1656406 8.715749e-08 -0.7086598 8.984848e-03
37 93183 PIGM -0.9816928 2.555046e-03 -0.7692141 4.412384e-02
38 50848 F11R -1.1850898 2.305841e-02 -1.7223230 6.574791e-04
39 257177 CFAP126 3.0271157 2.486380e-03 3.9977168 2.446772e-07
40 8490 RGS5 0.9871038 1.899387e-04 1.3713587 1.680433e-07
41 23127 COLGALT2 -0.7333432 9.324603e-03 -0.8582788 3.560033e-03
42 116496 NIBAN1 -1.0446381 1.625792e-02 -1.4691925 2.041723e-03
43 51377 UCHL5 1.5905726 4.556549e-02 1.8141245 3.961369e-02
44 23612 PHLDA3 -0.6288566 4.159805e-02 -0.9118039 1.552424e-02
45 89796 NAV1 0.8887002 5.271256e-03 0.7948131 1.376176e-02
46 134 ADORA1 -1.3185997 4.144645e-03 -1.4114659 5.096005e-03
47 1116 CHI3L1 -2.6115024 1.208695e-02 -2.1674289 2.611588e-02
48 947 CD34 -2.5431290 2.950214e-02 -3.8219667 1.470914e-03
49 50486 G0S2 -2.4260288 2.325505e-16 -3.0634207 1.068292e-20
50 7044 LEFTY2 -2.0064246 2.032404e-05 -3.3736191 3.771351e-10
51 56776 FMN2 2.1449606 4.807079e-06 2.1149674 2.037015e-05
52 1316 KLF6 1.1972400 2.196591e-08 1.1330613 1.624593e-04
53 2665 GDI2 3.1800776 3.018173e-02 3.2355077 3.509061e-02
54 7431 VIM 1.0615888 2.932357e-11 0.9328584 1.892918e-07
55 10529 NEBL 1.1141860 2.985345e-04 1.6529758 1.147439e-06
56 22852 ANKRD26 0.9005820 4.181479e-02 1.1633644 9.679134e-03
57 118738 ZNF488 -1.5589308 7.028558e-04 -1.6930972 1.044668e-03
58 8505 PARG 2.2988292 2.298951e-03 2.6818767 9.899451e-04
59 84159 ARID5B 1.4561376 1.818833e-15 1.6019328 7.389890e-10
60 221037 JMJD1C 1.1413654 4.873134e-02 1.4571420 3.007831e-02
61 5660 PSAP -0.5436960 1.899387e-04 -0.5930284 4.436193e-03
62 54541 DDIT4 -1.8999569 1.279542e-18 -1.5617952 1.558459e-10
63 25961 NUDT13 6.8227528 7.120887e-03 6.8823473 7.089486e-03
64 5328 PLAU -1.8051485 1.424238e-07 -2.4161307 3.088200e-15
65 142891 SAMD8 1.2167269 3.807863e-02 1.4376791 7.264359e-03
66 27063 ANKRD1 -2.1195362 4.474603e-05 -2.4006620 1.471437e-04
67 1592 CYP26A1 -4.8596134 6.312146e-11 -2.4281066 4.992770e-04
68 9124 PDLIM1 -0.5229078 5.392838e-03 -1.3111088 1.811159e-10
69 10580 SORBS1 0.8916824 4.398108e-02 1.3113528 2.727619e-03
70 9849 ZNF518A 1.5401151 2.562268e-02 1.7890637 1.468074e-02
71 84171 LOXL4 -1.1962768 3.718183e-02 -2.3242991 5.587152e-06
72 3257 HPS1 -1.1398465 7.811903e-04 -0.9794147 1.688600e-02
73 9748 SLK 1.2409666 7.280947e-03 1.4740585 1.442922e-03
74 120 ADD3 1.0270899 2.295963e-05 1.1101210 1.904779e-03
75 9126 SMC3 0.8299034 3.179656e-02 1.0698870 1.476370e-02
76 3983 ABLIM1 0.8058028 6.818597e-04 0.7933721 2.654188e-02
77 975 CD81 -0.5212097 5.881688e-03 -0.7685199 1.330742e-04
78 283298 OLFML1 -3.1263200 7.831127e-04 -3.0036699 7.665164e-07
79 7003 TEAD1 1.1780833 9.794695e-06 1.6239063 9.020228e-09
80 10418 SPON1 2.1741508 6.755551e-15 2.5590703 1.068292e-20
81 100126784 <NA> -1.4015044 1.901541e-02 -1.4478429 1.853502e-02
82 24147 FJX1 1.1144123 6.120134e-09 0.8760488 1.735644e-04
83 90139 TSPAN18 -1.8384241 4.046303e-04 -2.3054290 7.972248e-06
84 90993 CREB3L1 -1.2735232 6.210652e-03 -1.2936317 1.366194e-02
85 55626 AMBRA1 -0.9927751 4.895349e-02 -1.2127677 3.509061e-02
86 79026 AHNAK 0.8884194 4.878484e-04 1.0278375 5.499695e-04
87 256364 EML3 -1.3784786 2.166718e-02 -1.4281668 1.640363e-02
88 6094 ROM1 -0.9464862 4.359757e-02 -1.8271874 9.460667e-04
89 6520 SLC3A2 -0.6010128 1.136755e-02 -0.8513390 3.004790e-03
90 5920 PLAAT4 -1.5864254 2.077763e-16 -1.6401471 4.845336e-11
91 11145 PLAAT3 -0.7196691 4.056531e-05 -0.7963243 9.134792e-03
92 26472 PPP1R14B -0.9247206 1.650139e-02 -1.2102668 2.526266e-03
93 4054 LTBP3 -0.8540564 2.105743e-03 -1.2806071 3.385228e-04
94 80227 PAAF1 -1.1675713 1.599730e-02 -1.6211909 6.014922e-03
95 871 SERPINH1 -0.4822611 3.565226e-02 -0.9819086 2.716955e-05
96 5058 PAK1 -0.7554188 1.835342e-02 -0.8378435 5.934077e-03
97 60494 CCDC81 2.2805826 4.294546e-02 2.7535993 2.388664e-03
98 120114 FAT3 1.0765492 5.326777e-05 1.6843150 5.489214e-06
99 79659 DYNC2H1 1.1983193 7.500672e-03 1.4897946 4.504253e-03
100 2893 GRIA4 1.5448086 5.843233e-03 2.3573803 3.549778e-06
101 7070 THY1 -1.9515256 8.943879e-06 -1.9971484 2.588397e-05
102 50937 CDON -2.1978705 8.412397e-05 -2.8331210 4.769691e-07
103 84623 KIRREL3 -2.0006115 3.381142e-03 -2.0826278 3.256629e-02
104 334 APLP2 -0.6596621 5.592386e-05 -0.6041049 8.866753e-03
105 65125 WNK1 0.6675285 3.574424e-02 0.8616293 4.456096e-02
106 2597 GAPDH -0.5248687 2.136306e-02 -0.6199905 7.119239e-03
107 7167 TPI1 -1.1268297 3.464276e-05 -1.1051538 2.591956e-04
108 9746 CLSTN3 -1.1395614 5.051479e-03 -1.3509561 1.148789e-03
109 2 A2M 1.3834893 3.889229e-09 1.2866302 2.710622e-05
110 2012 EMP1 1.6013894 8.618402e-05 2.0049140 2.388623e-06
111 11228 RASSF8 1.3770183 2.991154e-02 1.5437649 1.195964e-02
112 636 BICD1 0.7723611 4.502745e-02 0.9248198 3.104741e-02
113 440097 DBX2 4.1236720 2.761389e-02 4.2129865 1.783039e-02
114 1280 COL2A1 3.9736137 2.121007e-03 4.0362936 2.637820e-04
115 51474 LIMA1 1.3522772 3.141070e-10 1.3088797 1.029063e-04
116 3164 NR4A1 2.3621346 3.107017e-03 2.5075572 1.471107e-03
117 3489 IGFBP6 -3.1684362 9.884164e-35 -3.6650546 5.021148e-35
118 3679 ITGA7 -0.7720557 3.551682e-02 -0.7646140 3.652514e-02
119 1606 DGKA -1.1418467 2.520844e-03 -2.0640839 6.920572e-07
120 6866 TAC3 -4.4018343 9.008210e-05 -2.2010970 5.337774e-05
121 1019 CDK4 -0.8535941 1.868937e-02 -0.7184192 2.076911e-02
122 10576 CCT2 0.5185069 2.504219e-02 0.7208397 5.157943e-03
123 6857 SYT1 1.3579287 1.590906e-04 1.5130491 2.820452e-05
124 160335 TMTC2 -1.2322975 2.485890e-02 -1.4552058 4.724671e-03
125 84125 LRRIQ1 1.6190068 1.882625e-02 2.1757057 2.192423e-03
126 80184 CEP290 1.3954645 6.795816e-04 1.3688197 1.084185e-02
127 1848 DUSP6 1.2880900 4.095711e-07 1.3562059 3.285592e-05
128 429 ASCL1 -1.2001440 2.092578e-02 -1.5098689 1.537816e-03
129 51559 NT5DC3 -1.3971847 1.319610e-02 -2.4564489 8.004114e-05
130 4938 OAS1 -6.0655526 3.961964e-05 -4.3761550 7.202309e-04
131 9921 RNF10 -0.6753467 1.710313e-03 -0.6977640 1.657455e-02
132 9612 NCOR2 -0.7709235 2.448638e-04 -1.0968871 1.174579e-04
133 53342 IL17D 1.4271867 3.117730e-02 1.4868968 3.660421e-02
134 26524 LATS2 1.9738166 2.631665e-12 2.1051688 1.177164e-11
135 55213 RCBTB1 2.4026672 4.745192e-02 2.5534468 3.778203e-02
136 10150 MBNL2 1.1988580 8.025352e-07 1.0849575 6.803360e-04
137 1948 EFNB2 0.8930437 1.142266e-04 1.0379934 2.191155e-04
138 1284 COL4A2 -0.7522418 7.234623e-06 -0.8132902 1.934513e-03
139 9985 REC8 -0.8811120 3.790527e-05 -0.9146541 5.019194e-04
140 54813 KLHL28 1.5878165 4.359757e-02 1.8279565 1.957397e-02
141 10979 FERMT2 0.8668750 2.009076e-05 0.8587077 5.216472e-04
142 652 BMP4 5.2239530 1.034773e-06 4.5812032 3.787086e-04
143 57161 PELI2 1.0098966 1.279683e-02 1.8089457 1.415149e-04
144 5015 OTX2 -4.1724443 6.014753e-04 -7.7513998 6.780151e-06
145 3306 HSPA2 1.1031974 3.816204e-02 1.7831662 9.821393e-06
146 87 ACTN1 -0.4715602 3.230139e-03 -0.6530761 7.189627e-03
147 89932 PAPLN -2.0434982 3.788123e-03 -2.4396537 1.240604e-03
148 91748 MIDEAS 1.8437688 2.596062e-04 1.4097996 3.078606e-02
149 91750 LIN52 7.3586806 7.558117e-04 6.7929895 3.676167e-02
150 7043 TGFB3 -2.3806142 3.579420e-04 -1.6424905 1.573491e-03
151 1734 DIO2 -2.8787481 3.934404e-03 -2.8579185 2.733301e-04
152 10516 FBLN5 2.1203131 2.339444e-21 2.6672653 1.068292e-20
153 4707 NDUFB1 -0.6371749 1.613544e-02 -0.6880549 3.741672e-03
154 90050 FAM181A 0.7026767 3.257981e-02 0.8317661 1.463922e-02
155 83982 IFI27L2 -0.7858180 3.788123e-03 -0.9554009 8.663085e-03
156 12 SERPINA3 -1.0928097 5.435726e-07 -2.1659703 5.383703e-08
157 317762 CCDC85C -1.1415379 1.960104e-04 -0.9268366 2.085976e-02
158 7453 WARS1 -1.1137925 7.403891e-03 -1.1877257 3.840242e-04
159 1778 DYNC1H1 -0.8599978 3.644681e-05 -0.7127652 1.666965e-02
160 23186 RCOR1 1.5421005 3.312328e-02 1.7881154 8.984848e-03
161 7127 TNFAIP2 -2.0371750 1.086495e-23 -2.2230001 2.531007e-10
162 7057 THBS1 1.7083931 2.189633e-03 2.4116586 5.357471e-06
163 2628 GATM 1.6305164 6.988424e-10 1.7248976 2.529714e-09
164 80031 SEMA6D 0.9025637 4.007236e-02 1.1477096 1.054213e-02
165 55329 MNS1 1.0871432 2.213310e-02 1.3100351 2.385152e-02
166 51495 HACD3 0.6985591 1.459480e-03 0.7733559 2.137496e-02
167 5315 PKM -1.0650906 6.410302e-11 -1.4937767 7.643068e-14
168 80381 CD276 -1.0479212 6.731425e-05 -1.3305282 6.040921e-05
169 4016 LOXL1 -1.1951972 5.642609e-10 -2.1437608 7.617542e-11
170 64220 STRA6 -1.5558629 1.022286e-08 -2.0378849 6.066252e-12
171 55272 IMP3 -1.1975167 1.407190e-04 -0.8758051 1.843729e-02
172 49855 SCAPER 1.2631768 2.255850e-02 1.4056129 1.565269e-03
173 1512 CTSH -1.3914988 9.906341e-04 -1.8041644 1.989209e-03
174 64782 AEN -0.7877903 2.899926e-02 -1.0936403 7.484975e-03
175 56963 RGMA -0.9149298 3.998689e-04 -1.3495988 1.632725e-08
176 18 ABAT 0.6707712 2.489975e-02 0.9341745 1.155951e-03
177 2903 GRIN2A 2.7357003 1.585718e-05 2.5921878 2.355363e-04
178 2013 EMP2 0.8598325 1.422745e-02 1.6100264 3.292616e-08
179 26471 NUPR1 -1.4791543 1.934933e-04 -1.5364080 4.009143e-02
180 27324 TOX3 -0.8199818 1.147411e-03 -0.9382922 4.351330e-03
181 54947 LPCAT2 1.2308899 2.758666e-02 1.2647993 3.027623e-02
182 4502 MT2A -0.8447907 1.111632e-05 -0.7476092 1.124292e-02
183 4498 MT1JP 4.9581335 2.520844e-03 5.0603883 2.388664e-03
184 6236 RRAD -1.4864441 1.438928e-06 -1.6488085 2.342924e-04
185 10204 NUTF2 -0.6198159 3.725852e-02 -0.7917682 1.237731e-02
186 5699 PSMB10 -1.1418958 1.319610e-02 -1.0338106 2.336391e-02
187 6560 SLC12A4 -0.8375102 2.808440e-02 -1.2495190 1.618789e-03
188 54921 CHTF8 -0.8281115 4.812666e-02 -0.9228884 4.653449e-02
189 497190 CLEC18B;CLEC18C -1.8130441 3.442171e-04 -1.5604160 8.663085e-03
190 57687 VAT1L 1.2538154 6.312146e-11 1.3033702 1.342139e-07
191 4094 MAF 0.9101083 1.475176e-02 1.5246996 8.004114e-05
192 54758 KLHDC4 -1.8537784 3.582707e-02 -1.9647262 3.778203e-02
193 8140 SLC7A5 -0.7394130 1.181188e-03 -1.5340156 1.559598e-12
194 79007 DBNDD1 -2.0251699 5.263677e-08 -1.7206308 7.979702e-05
195 1107 CHD3 -1.1390686 4.908692e-03 -1.0562523 3.467720e-03
196 9423 NTN1 -0.7862733 3.658209e-03 -0.8149528 2.312917e-02
197 5376 PMP22 0.6967419 3.417416e-03 1.0885869 1.085864e-04
198 4642 MYO1D 6.4094460 4.045680e-02 7.1055708 2.424094e-03
199 124842 TMEM132E -3.1037068 7.029061e-17 -3.2230094 2.966224e-02
200 60681 FKBP10 -0.9052788 8.382146e-06 -1.2543964 4.746007e-06
201 4836 NMT1 -0.6023684 3.380518e-02 -0.9041986 2.130740e-02
202 113026 PLCD3 -0.7450877 3.417416e-03 -0.8652446 5.923701e-03
203 55073 <NA> 3.4669716 5.421452e-03 3.3104655 3.014578e-02
204 55163 PNPO -1.1046242 2.060921e-03 -1.2146003 2.741049e-02
205 4804 NGFR -1.3065387 6.110275e-03 -1.2489577 4.207207e-02
206 1655 DDX5 0.4717216 4.846293e-02 0.6097198 3.556703e-02
207 3759 KCNJ2 1.9727944 1.468127e-02 2.7100478 8.004114e-05
208 6662 SOX9 0.6457921 3.502880e-02 1.1646511 1.925466e-03
209 2232 FDXR -1.2120535 8.486818e-03 -1.8688867 3.757563e-05
210 51155 JPT1 -0.5536597 1.488710e-02 -0.7414208 6.932722e-03
211 91107 TRIM47 -2.0017119 1.075934e-10 -2.3827524 3.446834e-08
212 284184 NDUFAF8 -0.6706425 1.598148e-03 -0.8440136 8.203951e-04
213 348262 MCRIP1 -0.7523709 4.465562e-02 -1.0446589 1.688600e-02
214 5034 P4HB -0.4543854 2.142260e-02 -0.5078332 4.207207e-02
215 2194 FASN -0.7235800 1.515178e-02 -0.9360883 1.242792e-02
216 114876 OSBPL1A 1.3057841 2.104057e-06 1.5147002 9.821393e-06
217 361 AQP4 1.0557538 2.520844e-03 1.8390826 3.459074e-05
218 4645 MYO5B -1.4080417 2.652893e-02 -1.8051401 3.761031e-03
219 5596 MAPK4 2.3332187 2.443723e-04 2.6647468 4.371215e-07
220 23239 PHLPP1 0.8908202 3.012397e-04 1.2308454 8.577989e-07
221 596 BCL2 1.3281477 3.874023e-02 1.7517641 2.702571e-04
222 10272 FSTL3 -1.3480049 1.332733e-03 -1.5364226 6.616941e-04
223 10975 UQCR11 -0.4736007 2.662868e-02 -0.7172415 2.358518e-02
224 2872 MKNK2 -0.7362165 6.709186e-03 -1.0370836 9.246743e-04
225 4616 GADD45B 1.1974882 1.680947e-04 1.0593744 2.957840e-02
226 91039 DPP9 -1.4129726 3.788123e-03 -1.5745440 1.997669e-02
227 126328 NDUFA11 -0.6662896 6.487686e-03 -0.7165509 1.102890e-02
228 1264 CNN1 -1.0302346 1.249444e-04 -1.5088944 6.861112e-07
229 54858 PGPEP1 -1.1297057 1.742422e-04 -0.7930293 3.207166e-02
230 1054 CEBPG 1.7212404 1.399782e-02 1.9076082 2.737070e-03
231 826 CAPNS1 -0.7145712 7.969899e-03 -1.0190790 8.881903e-03
232 7538 ZFP36 2.7061502 1.417965e-07 1.9880521 8.007645e-03
233 126299 ZNF428 -0.5164540 2.950214e-02 -0.6152886 4.279761e-02
234 602 BCL3 -0.7562378 1.619694e-02 -1.3403036 4.705297e-04
235 27113 BBC3 -0.8478769 1.155051e-04 -1.2765873 2.010192e-08
236 51171 HSD17B14 -1.9347867 4.730163e-03 -2.2523362 1.515824e-03
237 581 BAX -1.1654632 7.626497e-07 -1.0925439 7.493213e-05
238 2109 ETFB -0.9106383 5.326777e-05 -0.9558306 5.797112e-03
239 84787 KMT5C -2.0110488 2.390716e-03 -1.3856015 3.824844e-02
240 9741 LAPTM4A -0.5104741 2.345858e-02 -0.8891868 5.337774e-05
241 6382 SDC1 -1.9426375 4.652286e-03 -2.7204541 4.015322e-04
242 5500 PPP1CB -0.5705857 2.790987e-03 -1.0125722 1.703389e-04
243 81606 LBH 1.1963265 1.735848e-03 1.1103523 1.590201e-02
244 51232 CRIM1 1.0857103 1.057254e-05 1.3469841 6.938661e-05
245 618 BCYRN1 -1.1601381 9.081175e-05 -1.1613696 7.819963e-06
246 6711 SPTBN1 0.6343670 2.060604e-02 0.8131394 3.613945e-03
247 55704 CCDC88A 0.6293154 4.667836e-02 0.8803134 2.015144e-02
248 400986 ANKRD36C 0.9929078 2.758666e-02 1.0779072 3.204514e-02
249 9486 CHST10 -0.9986909 5.914655e-04 -0.9125609 2.581052e-04
250 84417 ECRG4 1.0447776 1.017080e-02 1.1899241 1.140768e-02
251 3625 INHBB -0.9333406 6.110275e-03 -1.6182067 4.316925e-06
252 2995 GYPC -1.7890666 1.147888e-02 -1.5761938 1.546050e-02
253 56475 RPRM 4.3557549 9.194609e-08 4.7505184 9.777715e-10
254 129881 CFAP210 2.4441067 2.404514e-04 2.4975404 1.149701e-03
255 6741 SSB 1.0786495 2.929416e-03 1.0547010 4.889999e-03
256 1123 CHN1 0.6380941 3.012397e-04 0.9383991 2.164894e-03
257 151246 SGO2 1.9823543 4.571128e-02 1.9011537 4.412384e-02
258 10152 ABI2 0.9004568 4.318369e-04 1.0801353 6.525642e-05
259 8609 KLF7 1.3561272 5.532541e-09 1.1691658 5.337774e-05
260 10314 LANCL1 1.7968983 1.529627e-04 1.5778927 3.967121e-03
261 2066 ERBB4 2.4737815 1.139811e-03 3.7510411 1.559598e-12
262 79582 SPAG16 0.6637017 4.255476e-02 0.8716435 3.661282e-03
263 3485 IGFBP2 -0.7785655 1.469109e-06 -0.8160109 1.066367e-02
264 3488 IGFBP5 -2.1399895 3.375354e-18 -2.2227097 4.196115e-14
265 5270 SERPINE2 -0.9375060 3.932808e-04 -0.7998009 2.723773e-02
266 3667 IRS1 1.2564193 1.613544e-02 1.3555660 1.161353e-02
267 92737 DNER 0.7296541 4.812564e-02 0.9400442 1.042755e-02
268 257407 C2orf72 1.6671501 5.102206e-03 1.9973846 1.250540e-04
269 3769 KCNJ13 2.6497393 1.223317e-03 3.3353468 1.119341e-06
270 2817 GPC1 -1.1264015 1.592620e-05 -0.9542791 2.424094e-03
271 4826 NNAT 2.7092363 1.981545e-08 3.2211410 4.568973e-11
272 84969 TOX2 -1.1107046 1.867304e-02 -1.6088850 1.333148e-02
273 55959 SULF2 -1.7097237 9.432701e-04 -2.0593723 2.754289e-03
274 655 BMP7 0.6516863 2.345858e-02 0.8062005 1.037066e-02
275 56937 PMEPA1 -0.7185488 4.427389e-04 -0.7973414 5.640513e-03
276 11047 ADRM1 -0.8361599 1.042566e-02 -1.0552800 1.143328e-03
277 140578 CHODL 1.5178684 3.765851e-05 1.6072225 2.116851e-05
278 10600 USP16 1.1221561 1.822845e-03 1.1420910 1.481960e-02
279 1827 RCAN1 1.1034291 1.388433e-08 1.3209653 7.276523e-10
280 7267 TTC3 0.5353325 3.730806e-02 0.7616492 1.911044e-02
281 5211 PFKL -0.8550956 8.824446e-03 -1.0213064 3.779815e-02
282 51586 MED15 -0.7463114 2.991154e-02 -0.7216400 4.343222e-02
283 6948 TCN2 -1.8365276 9.348294e-03 -1.8121841 2.914156e-04
284 7078 TIMP3 0.7308883 1.832688e-02 1.2375753 3.285592e-05
285 8542 APOL1 -3.6489390 1.529627e-04 -3.9693213 6.526148e-05
286 114904 C1QTNF6 -1.1670092 3.940409e-02 -1.2996292 1.390113e-02
287 23616 SH3BP1 -1.4965495 4.654776e-03 -2.8340251 4.746007e-06
288 11078 TRIOBP -0.4607248 3.710617e-02 -0.7691425 6.964023e-03
289 27350 APOBEC3C -2.7579760 4.807079e-06 -2.5020867 9.509294e-05
290 2847 MCHR1 -2.9047919 8.531027e-04 -3.0086149 1.168141e-03
291 2917 GRM7 6.8558864 9.504412e-03 7.2009222 2.874746e-03
292 100505641 FGD5-AS1 0.7406840 2.692463e-02 0.8929744 3.154993e-02
293 25907 TMEM158 -1.2392679 7.283377e-05 -1.2408605 2.786605e-04
294 79442 LRRC2 4.8032430 7.562671e-04 5.0152818 1.250080e-04
295 51246 SHISA5 -0.9021016 1.958157e-03 -0.9289792 6.641636e-04
296 57060 PCBP4 -1.0918143 1.277098e-02 -1.2812909 5.923701e-03
297 11344 TWF2 -0.8035449 6.297992e-03 -1.2698839 1.082406e-04
298 7086 TKT -0.9438676 2.130579e-06 -1.4724809 1.953998e-07
299 55079 FEZF2 -1.3819673 2.673481e-03 -1.3649965 9.117715e-04
300 23429 RYBP 1.1986580 1.731990e-02 1.3075760 9.873759e-03
301 253559 CADM2 1.6130357 2.521553e-03 2.0703621 1.683733e-04
302 56650 CLDND1 1.1424184 1.399319e-03 1.2241964 3.466028e-03
303 961 CD47 0.7496940 2.373138e-03 0.6692668 3.207166e-02
304 55081 IFT57 1.0677831 5.500106e-03 1.1381152 1.295695e-03
305 151887 CCDC80 0.6897242 7.479143e-03 0.8284530 3.603351e-04
306 2596 GAP43 -0.5860822 3.465204e-03 -0.6833783 6.643199e-03
307 54437 SEMA5B -0.6221855 1.882625e-02 -0.8301337 3.190479e-02
308 79858 NEK11 1.5590867 3.444604e-02 2.4213715 1.760994e-06
309 7018 TF -1.8927271 1.818833e-15 -1.0115731 6.828350e-04
310 51421 AMOTL2 1.2107443 6.806849e-04 1.4062631 1.646695e-04
311 5947 RBP1 -1.5865579 2.015005e-26 -1.2966017 1.225898e-07
312 5806 PTX3 1.8462867 7.199573e-06 2.0405618 6.343051e-09
313 10417 SPON2 -3.3609289 3.532954e-06 -3.7237338 1.118777e-07
314 57537 SORCS2 -0.9762227 3.486187e-02 -1.4798215 7.583063e-04
315 85460 ZNF518B 1.8047082 4.183023e-03 1.7601318 4.889999e-03
316 259282 BOD1L1 0.5201945 4.759144e-02 0.7616620 2.566650e-02
317 22998 LIMCH1 0.5964996 9.906341e-04 0.6136851 2.601096e-02
318 9508 ADAMTS3 1.5668308 2.722280e-02 1.8080564 1.307750e-02
319 79960 JADE1 1.2561816 4.257289e-03 1.0557625 3.961369e-02
320 57575 PCDH10 1.4842430 2.442048e-04 1.8355075 2.065426e-05
321 80854 SETD7 -0.8185458 4.900421e-02 -1.4305901 3.629990e-04
322 2549 GAB1 0.8933994 9.968826e-03 1.1763303 5.336430e-03
323 5188 GATB -0.9696811 1.128238e-02 -0.8148340 2.601096e-02
324 6307 MSMO1 0.8202208 3.063575e-03 0.9904135 4.008274e-04
325 23022 PALLD 0.8203477 3.343707e-03 0.6517112 4.026703e-02
326 3148 HMGB2 1.3902300 9.358605e-04 1.4964408 3.466028e-03
327 80014 WWC2 1.5077252 5.569873e-03 1.6337916 4.705297e-04
328 8470 SORBS2 0.7774946 3.028106e-04 0.6294174 2.385152e-02
329 2195 FAT1 0.8131067 5.051479e-03 1.3978566 2.702571e-04
330 23379 ICE1 1.0853409 3.117730e-02 1.3296719 3.778203e-02
331 134121 C5orf49 1.5327179 9.651516e-03 2.3053458 8.702439e-07
332 3157 HMGCS1 0.7391885 2.673481e-03 1.0037165 2.526266e-03
333 55914 ERBIN 0.8272980 1.835342e-02 1.4538577 3.718866e-04
334 3156 HMGCR 0.9055108 8.346949e-03 1.0600570 1.134561e-02
335 133746 JMY 0.9081683 2.177853e-02 1.1422575 7.616687e-03
336 7060 THBS4 2.3443647 6.885590e-06 2.3302850 6.274778e-04
337 7025 NR2F1 -0.8066261 6.795816e-04 -0.9269345 2.192423e-03
338 1105 CHD1 1.2242602 1.189098e-02 1.6028340 4.419966e-03
339 1946 EFNA5 2.4408090 3.090004e-05 2.4804390 6.648397e-05
340 324 APC 1.0410874 1.111810e-03 1.5315701 2.297585e-04
341 1456 CSNK1G3 1.4732785 7.318522e-03 1.8832219 2.430956e-03
342 57507 ZNF608 1.1772239 4.419816e-02 1.2535980 1.077182e-02
343 401207 C5orf63 -0.7152955 1.241150e-02 -0.9131935 2.192423e-03
344 84466 MEGF10 1.8092252 3.544644e-06 1.1733286 3.522841e-02
345 8572 PDLIM4 -0.8042731 2.592049e-02 -1.0260482 1.295695e-03
346 27089 UQCRQ -0.5361123 3.637772e-03 -0.6639010 3.420727e-02
347 9547 CXCL14 -1.8687526 1.770465e-32 -2.5656011 1.495936e-39
348 6695 SPOCK1 -1.6682759 7.720578e-06 -2.7112224 2.012757e-12
349 1958 EGR1 2.4598152 1.795437e-04 2.5466942 5.669999e-04
350 51247 PAIP2 0.8721594 5.841692e-04 0.8662296 1.326221e-02
351 1809 DPYSL3 -0.9696835 8.589897e-08 -0.9035729 1.338876e-05
352 11346 SYNPO -1.5238124 1.211177e-09 -1.7570515 3.664102e-08
353 55568 GALNT10 0.7896552 5.448953e-03 0.7079355 3.116160e-02
354 26999 CYFIP2 -0.7511372 6.961383e-03 -0.7315871 3.358754e-02
355 9232 PTTG1 -0.9989437 4.952622e-03 -0.9646327 5.266939e-03
356 23286 WWC1 0.8085005 1.452971e-02 1.0887096 4.705297e-04
357 1843 DUSP1 2.5268616 3.718090e-03 2.5402099 3.307831e-03
358 8614 STC2 -1.8578146 4.206285e-04 -1.8523512 2.637820e-04
359 84851 TRIM52 6.3617317 4.417802e-03 6.9732875 4.166404e-04
360 221749 PXDC1 0.9011447 7.283377e-05 1.1085754 1.077860e-04
361 4739 NEDD9 1.2738068 9.794695e-06 1.0696236 3.707999e-03
362 84830 ADTRP 1.6656204 1.378988e-02 2.3137138 1.161371e-05
363 3400 ID4 1.5750061 1.104334e-15 1.5869603 3.771351e-10
364 10537 UBD -2.0873239 7.455735e-08 -2.8005498 6.241412e-09
365 3105 HLA-A -1.5239966 4.838244e-06 -0.9968184 2.108854e-02
366 3106 HLA-B -1.0357628 8.563055e-07 -1.0585646 8.852170e-04
367 6892 TAPBP -0.8871305 4.199675e-05 -1.0759009 2.204245e-04
368 1026 CDKN1A -0.8316808 8.384613e-08 -1.5054481 9.779892e-15
369 2739 GLO1 0.6928464 3.131227e-02 0.7675737 4.451486e-02
370 4188 MDFI -2.2086189 2.257366e-09 -1.7774680 5.591040e-05
371 5429 POLH -1.1784592 4.417802e-03 -1.1081187 9.804193e-03
372 667 DST 1.2629742 1.424238e-07 1.9534245 4.726491e-02
373 79940 LINC00472 1.4444928 3.865318e-02 2.8962830 3.076670e-07
374 1915 EEF1A1 0.6271181 2.887853e-03 0.6368240 4.653449e-02
375 100133941 CD24 1.1934937 1.144762e-06 1.3240155 1.981373e-07
376 285755 PPIL6 1.8827377 8.169943e-03 2.0478083 2.310070e-03
377 3910 LAMA4 0.7188565 3.874023e-02 0.8198526 4.916032e-02
378 2697 GJA1 1.3765753 7.105437e-23 1.4624145 1.582151e-14
379 154214 RNF217 2.4102950 1.268124e-05 2.1900125 1.007178e-03
380 1490 CCN2 2.2575437 1.770465e-32 2.0256712 1.068292e-20
381 51696 HECA 2.3673205 1.842907e-02 2.2035454 4.158057e-02
382 6648 SOD2 -0.7015835 7.479143e-03 -0.8804546 7.841559e-03
383 168090 C6orf118 6.2591937 3.551133e-04 7.0656738 2.275590e-06
384 2115 ETV1 1.3607620 2.074310e-06 1.5885440 8.567961e-12
385 6424 SFRP4 -1.0873687 1.224412e-10 -1.6506217 3.349551e-14
386 10652 YKT6 -0.6105490 3.925318e-02 -0.7509839 2.808502e-02
387 3486 IGFBP3 1.9252602 8.715749e-08 1.5836464 2.355363e-04
388 51119 SBDS 1.1784616 5.957958e-03 1.0523699 3.961369e-02
389 64409 GALNT17 -1.1073150 3.644681e-05 -1.8657379 2.178280e-08
390 155370 <NA> 1.7221992 1.636217e-02 1.5972194 4.067875e-02
391 3984 LIMK1 -0.7406690 4.299917e-02 -1.4173691 3.193715e-04
392 9988 DMTF1 2.7844122 1.402480e-02 3.2086119 9.702197e-04
393 1021 CDK6 0.7718891 4.359757e-02 1.1586428 1.755742e-02
394 7980 TFPI2 0.8418426 5.914655e-04 1.0491806 7.493213e-05
395 2791 GNG11 0.8358781 1.628460e-02 0.8496367 2.966224e-02
396 222865 TMEM130 -2.3271890 8.320523e-04 -2.1581089 1.783039e-02
397 10095 ARPC1B -1.1716736 2.673919e-03 -1.0093216 1.085725e-02
398 79690 GAL3ST4 -1.5038820 4.590741e-05 -1.3642001 1.126403e-03
399 5054 SERPINE1 1.9484639 4.805049e-07 1.5218928 1.995334e-03
400 1174 AP1S1 -0.8686414 3.245913e-02 -1.0947152 4.889999e-03
401 136227 COL26A1 0.9856836 1.613544e-02 1.2308276 3.746490e-03
402 10234 LRRC17 -2.8352184 2.959956e-02 -5.0413958 3.788070e-09
403 50640 PNPLA8 1.7965209 5.004492e-03 1.6304588 2.346602e-02
404 858 CAV2 -1.4532955 4.417802e-03 -2.0232878 1.218165e-03
405 2861 GPR37 2.9270589 2.608155e-12 3.3869762 4.153477e-14
406 7328 UBE2H -0.4539210 2.866134e-02 -0.7003310 3.238744e-03
407 4708 NDUFB2 -0.7818392 2.026161e-03 -0.8137652 1.922160e-02
408 26047 CNTNAP2 -2.9235371 3.690939e-04 -2.7690271 3.811547e-03
409 155066 ATP6V0E2 -0.6425637 1.021301e-02 -1.0316768 5.966463e-05
410 51667 NUB1 1.2120480 2.190974e-02 1.4286863 2.490482e-02
411 349136 WDR86 -0.6793698 1.094448e-02 -0.8780693 9.830944e-03
412 10395 DLC1 1.7700732 1.230399e-10 1.5814496 2.266267e-07
413 57509 MTUS1 1.3279827 3.971037e-02 1.7515064 2.029801e-03
414 5108 PCM1 0.6296563 2.613782e-02 1.0944067 3.718866e-04
415 4747 NEFL -2.4697340 3.858818e-07 -2.4268546 2.427204e-06
416 1808 DPYSL2 0.8712415 1.424238e-07 0.9222317 5.410490e-06
417 81551 STMN4 -2.1760474 5.188815e-11 -2.5255769 3.088200e-15
418 80139 ZNF703 -1.6318203 1.459480e-03 -1.9408008 1.530981e-04
419 80223 RAB11FIP1 -1.8514669 1.883047e-04 -2.1552343 6.945104e-04
420 5327 PLAT -0.9426855 9.155802e-05 -1.2287574 6.636739e-06
421 9821 RB1CC1 1.6078703 4.349835e-06 1.3805131 1.902604e-02
422 286183 NKAIN3 -0.9164839 1.569422e-05 -0.9003674 1.843729e-02
423 8836 GGH -0.8820527 5.055733e-04 -1.3403599 1.751109e-04
424 83690 CRISPLD1 1.5438934 1.386708e-16 1.0629238 5.790464e-05
425 51101 ZC2HC1A 0.9197911 4.087158e-02 1.0266947 4.685597e-02
426 6383 SDC2 1.0676893 2.679821e-07 0.9792857 2.023716e-04
427 25897 RNF19A 1.1085351 8.624300e-03 1.3082186 1.528592e-02
428 114907 FBXO32 0.8178647 3.430723e-07 0.5884895 4.679822e-02
429 157638 LRATD2 0.8171015 2.592411e-03 0.7465006 1.859717e-02
430 10655 DMRT2 -7.3194679 1.427260e-03 -4.9765653 2.628093e-02
431 8777 MPDZ 0.9438101 3.487332e-02 1.3112202 5.732331e-04
432 11168 PSIP1 0.8441563 1.417117e-03 1.0080687 3.467720e-03
433 92949 ADAMTSL1 -2.4743892 2.404514e-04 -2.5083493 1.480496e-03
434 1029 CDKN2A -1.0698981 2.693797e-03 -1.0809118 5.318513e-03
435 2683 B4GALT1 -0.8050145 1.786092e-02 -1.5559157 1.731370e-05
436 23137 SMC5 0.8680319 2.565594e-02 1.0374083 4.283932e-02
437 687 KLF9 1.3492087 2.007537e-07 1.5378853 5.096218e-07
438 5727 PTCH1 1.9853072 5.970156e-03 1.9617776 5.954834e-03
439 8013 NR4A3 2.4904087 6.908980e-06 3.1746652 1.196718e-09
440 54566 EPB41L4B 1.8852877 4.469557e-05 1.8231148 8.637070e-05
441 401548 SNX30 2.9447858 7.197629e-03 2.6829839 3.424548e-02
442 5998 RGS3 -0.5840860 4.877383e-02 -0.7715307 1.638896e-02
443 399665 FAM102A -0.7378701 1.847333e-02 -0.7969264 2.473596e-02
444 80142 PTGES2 -1.1690185 4.957142e-02 -1.3531726 1.243737e-02
445 5524 PTPA -0.7293439 3.510046e-02 -0.9508710 1.700948e-03
446 389792 IER5L -0.7284427 9.413223e-03 -1.1608257 1.250080e-04
447 84814 PLPP7 -6.8009388 3.832155e-02 -6.7539332 4.025168e-02
448 1289 COL5A1 -1.8804761 3.712439e-07 -2.0532414 6.814519e-05
449 29085 PHPT1 -0.7767434 3.563725e-03 -0.6540889 3.522841e-02
450 5730 PTGDS -1.5236081 2.334225e-07 -1.4155954 3.718866e-04
451 4571 <NA> -1.2077125 1.926212e-03 -1.1240600 1.905700e-03
452 1756 DMD 0.8880564 9.122453e-03 1.4276635 3.041793e-04
453 83604 TMEM47 1.7104306 6.312146e-11 1.8614711 1.606743e-09
454 7317 UBA1 -0.5390180 2.867141e-02 -0.7015361 1.485338e-02
455 5127 CDK16 -0.8688021 6.334141e-03 -0.8488377 3.241757e-02
456 83692 CD99L2 -0.8252556 4.595109e-03 -0.7957799 1.366194e-02
457 1069 CETN2 0.5259222 2.712703e-02 0.8316609 2.042964e-04
458 633 BGN -1.4153441 1.019612e-02 -2.3268395 8.242040e-07
459 537 ATP6AP1 -1.1433512 8.346232e-05 -0.9590374 4.786404e-02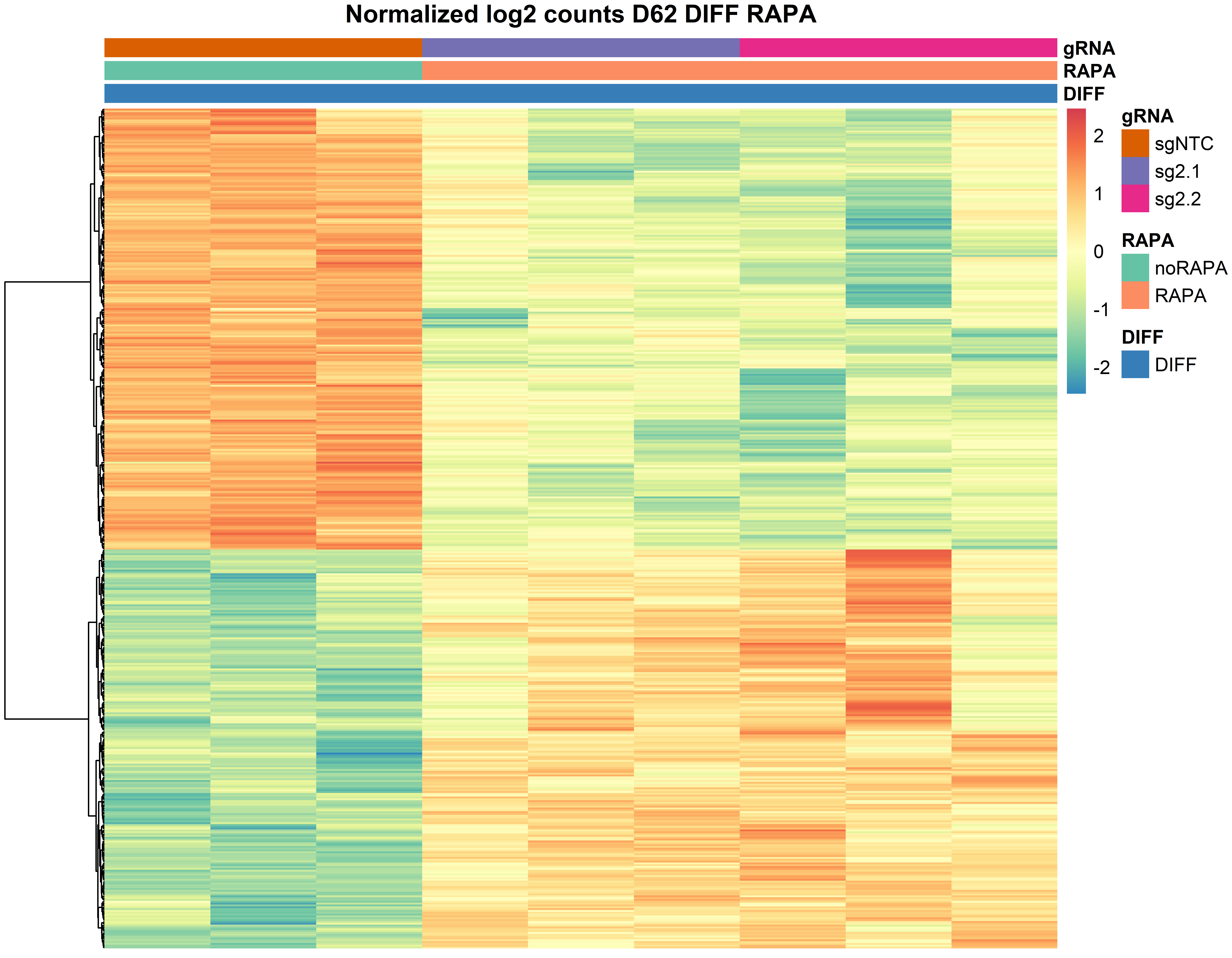
Warning: Removed 1 rows containing missing values (`geom_point()`).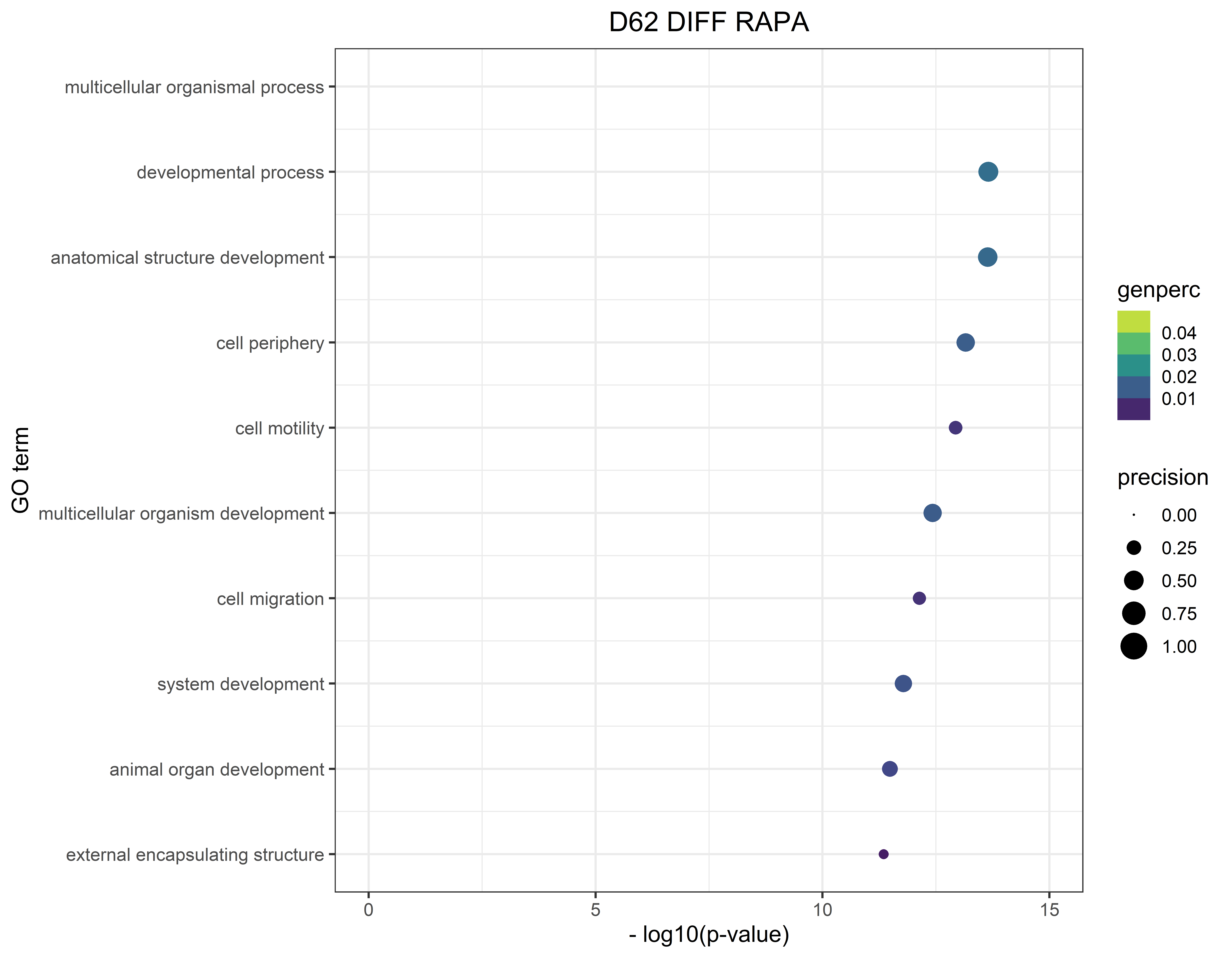
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.4756215 1.021464590 2.078115 3.274835e-02
02_TSC2_KO_Grabole_2016 1.6641362 1.373571755 2.014742 1.359647e-07
03_TSC2_Grabole-2016 1.8316335 1.504188246 2.236420 5.229449e-10
04_TSC2-tuber-Martin2017 4.2722382 2.233978893 7.593020 1.934504e-05
05_TSC2-SEN_SEGA-Martin2017 1.9827408 1.578703793 2.474985 6.044251e-09
06_all_EpilepsyGene 1.2843779 0.719840991 2.138225 3.102380e-01
07_high_conf_EpilepsyGene 1.4764095 0.528256524 3.335901 3.120259e-01
08_ASD_Epilepsy_EpilepsyGene 1.5847430 0.566201243 3.588209 2.885074e-01
09_Epi25_dataset 1.0713602 0.215628905 3.264358 7.598994e-01
10_SFARI_all 1.9413348 1.059688145 3.305647 1.853638e-02
11_SFARI_score4plus 1.7164023 0.802849809 3.264524 9.565232e-02
12_DeRubeis_RDNV 0.3646461 0.009100953 2.101845 5.260007e-01
13_Iossifov_RDNV 1.3104319 0.805102804 2.032759 2.111285e-01
14_FMRP_Darnell 1.2372069 0.823887776 1.798561 2.546266e-01
15_Voineagu_M12 0.9540530 0.468206892 1.746123 1.000000e+00
16_Voineagu_M16 3.0381129 2.003706438 4.466527 4.122801e-07
17_ID_genes_Neel 1.0199201 0.500220768 1.868455 8.755363e-01
18_ID_genes_pinto2014 0.4898401 0.131782214 1.279836 1.995030e-01
19_Fromer_SCZ_FunDN 1.1068666 0.644145572 1.789726 6.100315e-01
20_Cocchi_Pruning 4.9939093 2.525076123 9.136560 8.636787e-06
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.38907924 0.18767437 6.549671e-01
02_TSC2_KO_Grabole_2016 0.50930622 0.09790395 2.719295e-06
03_TSC2_Grabole-2016 0.60520822 0.10048716 1.045890e-08
04_TSC2-tuber-Martin2017 1.45213787 0.33079266 3.869008e-04
05_TSC2-SEN_SEGA-Martin2017 0.68448014 0.11626327 1.208850e-07
06_all_EpilepsyGene 0.25027444 0.29540785 1.000000e+00
07_high_conf_EpilepsyGene 0.38961313 0.52438082 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene 0.46042222 0.52511629 1.000000e+00
09_Epi25_dataset 0.06892908 0.81792115 1.000000e+00
10_SFARI_all 0.66337578 0.30887812 3.707276e-01
11_SFARI_score4plus 0.54023043 0.38766227 1.000000e+00
12_DeRubeis_RDNV -1.00882792 1.88293278 1.000000e+00
13_Iossifov_RDNV 0.27035679 0.24854189 1.000000e+00
14_FMRP_Darnell 0.21285633 0.20743739 1.000000e+00
15_Voineagu_M12 -0.04703610 0.36316781 1.000000e+00
16_Voineagu_M16 1.11123658 0.21236627 8.245603e-06
17_ID_genes_Neel 0.01972424 0.36348469 1.000000e+00
18_ID_genes_pinto2014 -0.71367624 0.66986142 1.000000e+00
19_Fromer_SCZ_FunDN 0.10153311 0.27620594 1.000000e+00
20_Cocchi_Pruning 1.60821903 0.34793256 1.727357e-04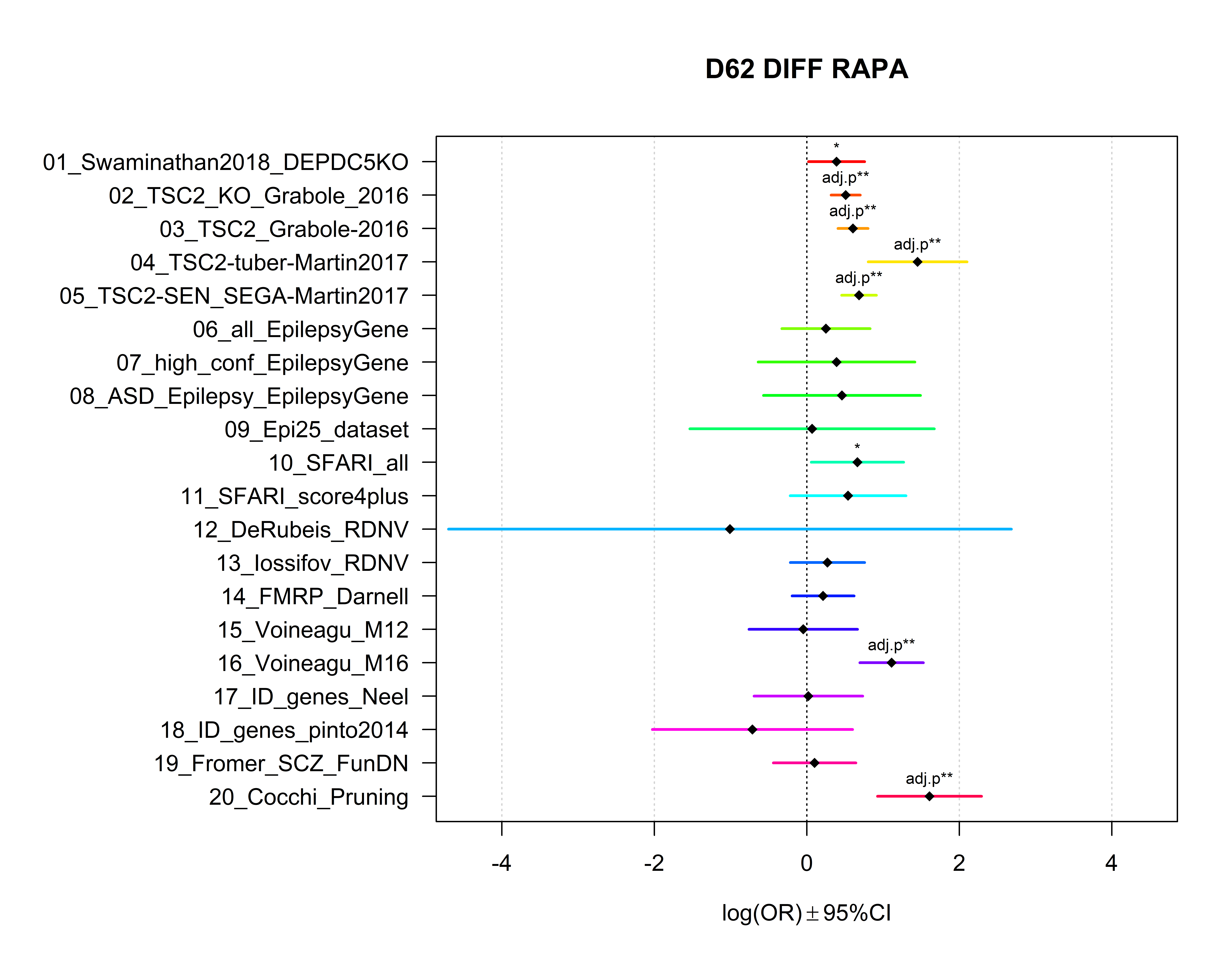
getlinmodoutput(targetdiff = "DIFF",targetrapa = "RAPA", refset = "NTCwRAPA" ) entrezgene genname log2FC_2.1 fdr_2.1 log2FC_2.2 fdr_2.2
1 2048 EPHB2 -0.5844492 1.601499e-02 -0.9155074 1.354145e-03
2 2537 IFI6 -0.7010783 8.040745e-03 -0.7815160 3.132856e-02
3 7802 DNALI1 1.1551849 7.457999e-03 1.1123245 2.753143e-02
4 656 BMP8B -4.3696453 8.803629e-05 -4.1397593 5.459052e-04
5 84879 MFSD2A 1.2281235 1.965733e-06 1.7552161 1.122712e-12
6 5865 RAB3B 1.6493707 4.465200e-03 2.4877824 6.594001e-08
7 3491 CCN1 0.9183982 7.579197e-06 0.7784458 2.753143e-02
8 2633 GBP1 1.8933713 1.501465e-03 1.7672396 3.995640e-03
9 127003 CFAP276 1.6882724 2.839987e-04 2.4595315 1.003361e-07
10 10628 TXNIP -1.1219260 2.851055e-03 -1.1836796 6.295463e-03
11 63827 BCAN -1.1464871 8.299200e-07 -0.6896098 4.022170e-02
12 3766 KCNJ10 -1.2044622 1.373679e-04 -1.1287694 2.169938e-03
13 257177 CFAP126 2.8399531 2.717907e-04 3.8109236 2.735624e-10
14 8490 RGS5 0.9430361 1.786616e-03 1.3273510 4.495792e-06
15 55220 KLHDC8A 0.6951021 4.636355e-02 1.0864127 8.650462e-04
16 50486 G0S2 -2.1451872 2.585499e-18 -2.7830791 4.179116e-23
17 7044 LEFTY2 -1.1468367 1.385907e-02 -2.5118705 2.149008e-06
18 2786 GNG4 2.3436557 4.465200e-03 3.1398129 1.562584e-07
19 1959 EGR2 3.8846116 1.679040e-14 3.0315167 5.126766e-05
20 84858 ZNF503 1.8062680 1.709089e-06 1.2785794 2.197425e-02
21 2674 GFRA1 -1.7276673 7.540604e-05 -1.9909032 7.225396e-05
22 5654 HTRA1 0.9265121 2.105270e-05 0.9286148 1.419288e-03
23 10418 SPON1 2.9482577 9.690519e-30 3.3331993 1.789378e-38
24 203859 ANO5 2.3880007 3.103096e-03 2.7523800 7.231881e-05
25 4038 LRP4 -0.7480393 8.990926e-03 -1.0202640 7.912549e-03
26 374393 FAM111B 3.2891956 3.036275e-02 3.5420547 1.522831e-02
27 79026 AHNAK 0.6654714 4.143655e-02 0.8048769 3.201026e-02
28 254263 CNIH2 1.8776550 1.040622e-04 2.0847320 2.685876e-06
29 2 A2M 1.5190611 3.193582e-12 1.4222515 1.439940e-06
30 79365 BHLHE41 1.2106415 6.905593e-04 1.2313105 3.995640e-03
31 6857 SYT1 1.6683728 8.962647e-06 1.8237151 2.110425e-06
32 101929005 <NA> -6.9708042 4.418423e-04 -4.6029204 9.008603e-03
33 114795 TMEM132B -2.0423423 5.591399e-15 -1.4960637 4.846059e-06
34 688 KLF5 1.6293748 1.038077e-03 1.2405652 4.163597e-02
35 1948 EFNB2 0.9649580 6.728368e-05 1.1098983 2.145665e-04
36 23002 DAAM1 0.7549476 2.716145e-02 0.8319282 2.753143e-02
37 6252 RTN1 1.7634057 2.333495e-12 1.5536847 2.770348e-04
38 89932 PAPLN -1.5738666 4.299050e-02 -1.9698221 1.607762e-02
39 10516 FBLN5 2.0017182 1.323846e-25 2.5486282 4.687237e-21
40 26153 KIF26A 4.9439872 4.867672e-03 5.7558523 3.737899e-05
41 7057 THBS1 1.3670907 2.165183e-03 2.0696822 4.051997e-06
42 4756 NEO1 0.9731714 2.838263e-08 1.1440173 1.493514e-05
43 4916 NTRK3 1.5301080 4.829515e-08 1.3001181 1.848612e-04
44 2903 GRIN2A 2.6806855 3.126489e-05 2.5384700 4.359783e-04
45 729993 SHISA9 -3.8906884 6.900445e-22 -1.3772648 3.301516e-03
46 51704 GPRC5B -0.6965404 1.535995e-02 -0.7199939 4.400840e-02
47 5579 PRKCB -6.4499033 3.151642e-11 -2.7969992 1.322087e-02
48 999 CDH1 -4.5747249 3.016659e-16 -2.8895584 2.205613e-07
49 54768 HYDIN 3.1138005 3.384464e-07 3.4374344 7.224320e-10
50 794 CALB2 1.9731509 2.785777e-03 2.4708643 1.488766e-06
51 57687 VAT1L 1.0676611 3.379263e-09 1.1174032 9.580576e-06
52 5187 PER1 1.6172545 2.138032e-03 1.9663057 3.759972e-06
53 201161 CENPV 1.0741222 7.146064e-03 1.3592159 2.907086e-04
54 7153 TOP2A 0.8905880 1.892264e-03 0.9745793 6.768778e-03
55 10882 C1QL1 -1.2476382 5.159240e-06 -1.2873491 1.702279e-03
56 113026 PLCD3 -0.7139995 2.028433e-02 -0.8342760 3.007915e-02
57 4804 NGFR -2.2035302 1.218551e-10 -2.1448207 1.880375e-06
58 2302 FOXJ1 1.2519998 4.679308e-06 1.3465537 1.186135e-03
59 11322 TMC6 -4.5309138 4.774950e-03 -4.0969189 3.201026e-02
60 361 AQP4 2.5635878 2.241622e-18 3.3473029 1.073340e-16
61 5596 MAPK4 1.9452939 5.842815e-04 2.2767959 8.837296e-08
62 51046 ST8SIA3 2.3074095 2.785777e-03 2.6047875 1.109491e-03
63 90701 SEC11C -0.6557054 3.229834e-02 -0.7760037 4.348299e-02
64 404217 CTXN1 1.0654569 6.973826e-05 1.0074858 9.552659e-04
65 1995 ELAVL3 1.3858314 1.866195e-05 2.2025879 3.191590e-13
66 9518 GDF15 -0.7426862 4.629676e-02 -1.6933332 4.065644e-05
67 79784 MYH14 -2.3284457 1.838639e-02 -3.3398391 1.027542e-03
68 92749 DRC1 2.1683527 2.785777e-03 2.6726025 3.759972e-06
69 57628 DPP10 -5.2202074 3.664045e-08 -3.4139399 3.737899e-05
70 3625 INHBB -0.9767644 8.988301e-03 -1.6615968 4.344424e-06
71 2995 GYPC -2.3374886 4.877829e-06 -2.1257263 2.391014e-06
72 56475 RPRM 1.7750559 4.755493e-02 2.1695403 5.813931e-03
73 129881 CFAP210 2.3640626 1.471679e-03 2.4162686 4.995507e-03
74 1134 CHRNA1 -5.2042681 2.191095e-11 -8.5037878 4.066660e-08
75 8324 FZD7 -1.3257906 1.734580e-04 -1.0432077 2.405225e-02
76 8609 KLF7 1.0346818 1.544898e-05 0.8476982 1.161968e-02
77 3488 IGFBP5 -1.2071406 1.460991e-03 -1.2892386 3.417235e-03
78 5270 SERPINE2 -1.3844484 2.152519e-07 -1.2467958 4.212466e-04
79 92737 DNER 1.1950369 4.465200e-03 1.4052480 1.004895e-03
80 3769 KCNJ13 1.7547914 2.427283e-02 2.4387114 2.984745e-05
81 4826 NNAT 3.6877783 4.792925e-26 4.2006138 4.152309e-28
82 55959 SULF2 -1.4289876 1.064627e-02 -1.7780168 2.556965e-02
83 56245 C21orf62 1.0634763 4.472992e-05 0.8769423 2.790773e-02
84 116448 OLIG1 -1.1527437 5.652288e-03 -1.1581057 1.134540e-02
85 1827 RCAN1 0.7187131 2.320004e-03 0.9362363 1.732037e-04
86 3708 ITPR1 1.9492555 8.038676e-03 1.9058343 3.555590e-02
87 116135 LRRC3B 1.9151330 1.659171e-03 1.7449314 1.677395e-02
88 114884 OSBPL10 -1.9432227 4.481965e-03 -1.7342826 3.185883e-02
89 79442 LRRC2 3.4370870 2.321235e-03 3.6464221 2.340152e-04
90 64147 KIF9 1.5017571 1.147328e-04 2.1367445 5.934113e-08
91 11170 FAM107A 1.4033068 3.229834e-02 1.4541890 4.163597e-02
92 132204 SYNPR 6.8506004 1.926905e-04 6.6427886 2.311735e-04
93 1295 COL8A1 -4.5472935 6.661090e-03 -3.7402461 2.753143e-02
94 151887 CCDC80 0.7296925 1.713718e-02 0.8683484 1.526926e-03
95 7018 TF -3.7184564 1.721396e-66 -2.8373320 5.562076e-30
96 5947 RBP1 -1.0936355 5.253202e-07 -0.8037544 4.130282e-02
97 5806 PTX3 1.5664566 1.373679e-04 1.7610651 1.723679e-07
98 10417 SPON2 -2.4659561 1.262912e-02 -2.8343935 2.249885e-03
99 83888 FGFBP2 -1.9573467 8.949057e-03 -2.3586659 2.311735e-04
100 201780 SLC10A4 1.0935517 7.536448e-06 1.0277894 5.530668e-03
101 23284 ADGRL3 -3.3750033 7.055351e-11 -1.7636588 1.363084e-03
102 9508 ADAMTS3 2.2329199 3.951529e-04 2.4736437 2.238692e-04
103 2920 CXCL2 8.1486161 1.339906e-04 7.8070786 2.129180e-04
104 134121 C5orf49 1.9601335 1.873352e-04 2.7327902 2.311097e-10
105 8507 ENC1 0.9054972 2.172818e-02 1.0168645 2.863416e-02
106 1946 EFNA5 1.6668921 1.152721e-06 1.7049758 2.723816e-05
107 9547 CXCL14 -0.8082314 7.395235e-04 -1.5055991 1.936896e-09
108 6695 SPOCK1 -1.1952779 8.323687e-03 -2.2385261 6.594001e-08
109 1958 EGR1 2.4384032 1.075179e-34 2.5255163 1.444721e-18
110 1839 HBEGF 0.7097541 4.806870e-02 1.2133655 2.381940e-05
111 2878 GPX3 -1.3039233 4.367899e-08 -2.0968752 8.969947e-11
112 23286 WWC1 1.0146586 3.812164e-03 1.2949889 1.181825e-04
113 1843 DUSP1 1.9070214 3.084357e-02 1.9207454 2.742071e-02
114 51617 NSG2 6.1428862 3.114379e-03 6.8867674 3.737899e-05
115 10814 CPLX2 5.4609870 4.418423e-04 6.4008339 1.003361e-07
116 221749 PXDC1 0.6427621 5.651259e-03 0.8503611 7.732420e-03
117 84062 DTNBP1 -0.9345057 1.444723e-02 -1.0734617 7.721019e-03
118 3400 ID4 1.0495736 1.369734e-04 1.0614846 3.046886e-03
119 10590 SCGN 3.4188121 6.157139e-11 4.5848750 6.141301e-26
120 7021 TFAP2B -8.8135379 1.003263e-07 -3.8657719 2.550146e-04
121 285755 PPIL6 1.9286056 1.035531e-02 2.0936062 3.182026e-03
122 1490 CCN2 0.9951832 6.973826e-05 0.7633184 2.561547e-02
123 80129 CCDC170 2.1577217 4.467859e-02 2.8156107 8.392469e-03
124 10457 GPNMB -1.5288283 8.425953e-04 -2.1689390 9.152129e-08
125 4852 NPY 3.1519799 7.989936e-05 4.2202467 1.680493e-14
126 6424 SFRP4 -0.7828072 4.392521e-04 -1.3460638 1.315271e-07
127 81628 TSC22D4 -0.6172060 1.874653e-02 -0.7801785 2.996625e-02
128 5054 SERPINE1 1.6825892 1.397417e-08 1.2560134 3.185320e-03
129 2861 GPR37 1.3652118 2.321235e-03 1.8253186 1.260450e-04
130 10395 DLC1 1.6104766 1.234236e-09 1.4218990 2.244802e-06
131 1808 DPYSL2 1.0102488 6.534125e-08 1.0613397 3.433965e-06
132 23643 LY96 -2.5960407 2.229924e-03 -1.9524795 4.587400e-02
133 11075 STMN2 4.3095979 1.701127e-02 5.8526106 8.836740e-09
134 6383 SDC2 0.8742833 2.175923e-04 0.7858399 1.707420e-02
135 6674 SPAG1 2.0758505 2.785777e-03 2.0003649 1.134540e-02
136 10655 DMRT2 -8.1117706 8.962647e-06 -5.7290427 5.822932e-04
137 1029 CDKN2A -1.0367133 1.263131e-03 -1.0483347 4.380981e-03
138 687 KLF9 1.4638994 4.727652e-09 1.6524793 5.934113e-08
139 9568 GABBR2 2.3438085 4.465200e-03 2.2371217 2.897821e-03
140 54566 EPB41L4B -1.0946069 1.984354e-03 -1.1567214 4.914294e-04
141 1620 BRINP1 2.9326320 2.760538e-02 3.7284868 2.949459e-05
142 83543 AIF1L -0.9372391 1.138078e-03 -0.9069212 2.561547e-02
143 8021 NUP214 -0.8751811 3.911704e-03 -0.9871582 5.616330e-03
144 5730 PTGDS -2.0124717 4.957753e-20 -1.9036600 7.962779e-09
145 4129 MAOB 1.2821359 1.974426e-05 1.0076275 1.211226e-02
146 1641 DCX 3.7333185 9.772500e-19 5.0031534 1.764598e-51
147 1069 CETN2 0.6722015 1.072993e-02 0.9779653 9.712269e-05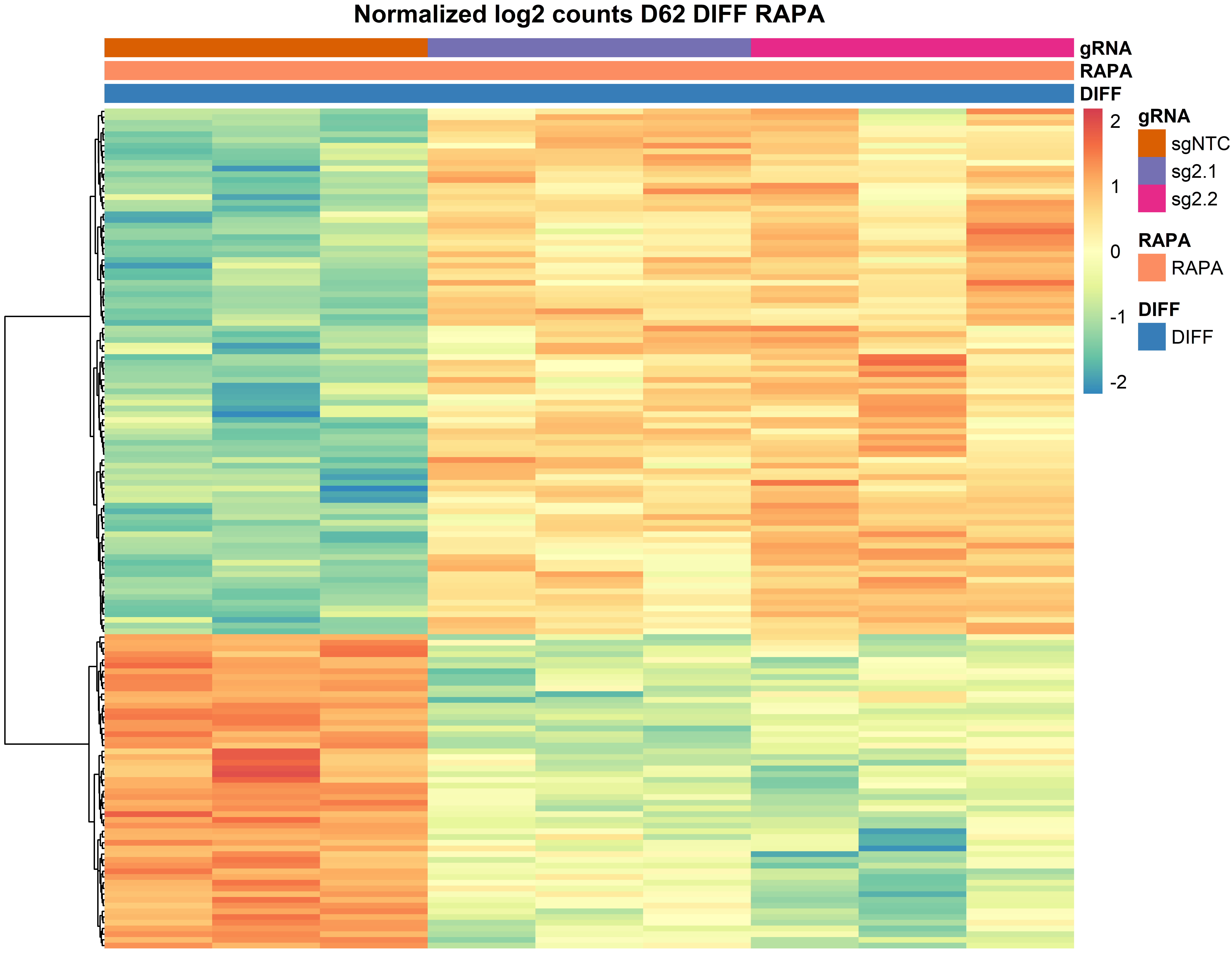
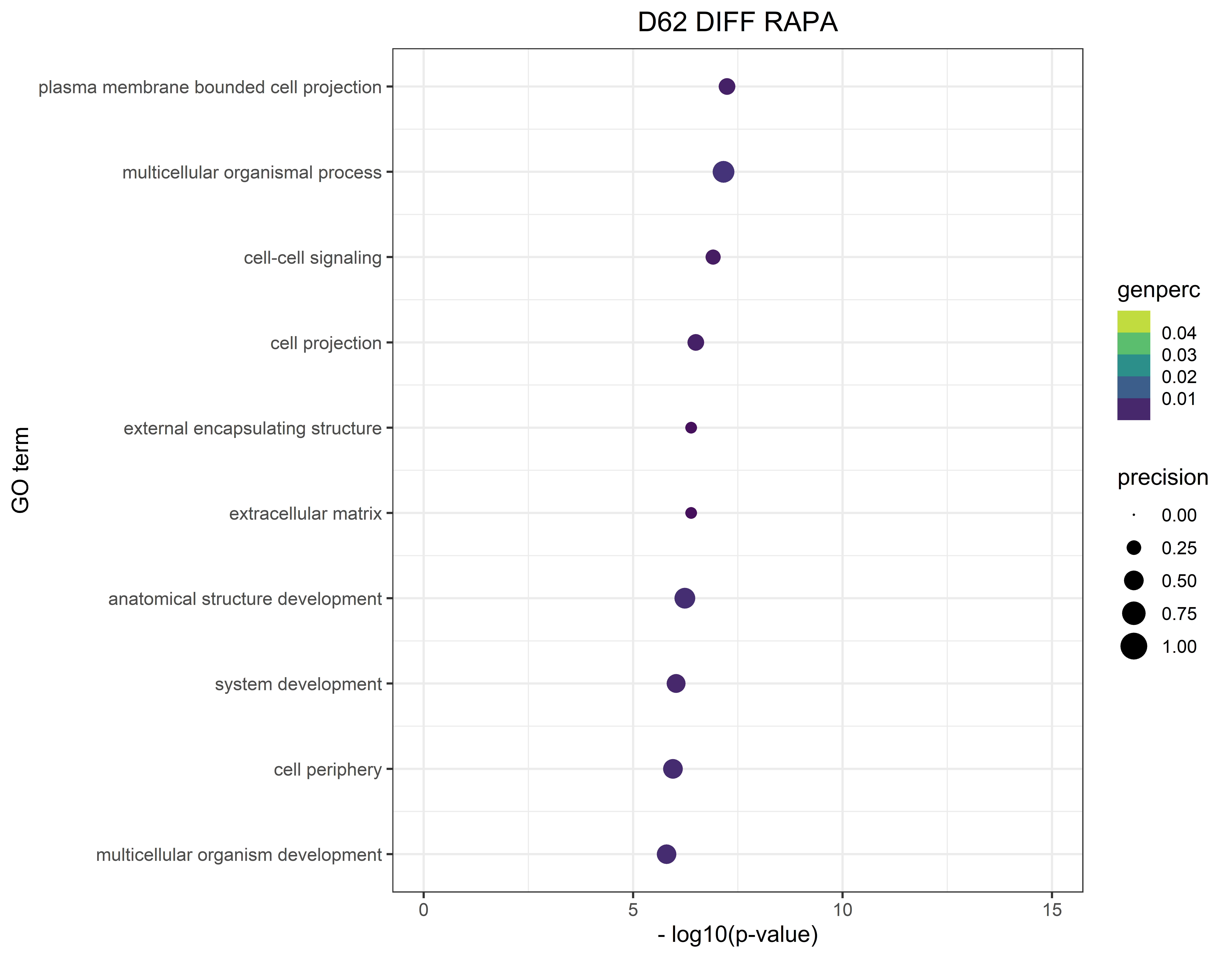
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.5710698 0.81157043 2.795221 1.528172e-01
02_TSC2_KO_Grabole_2016 2.1717985 1.54739180 3.053726 4.324330e-06
03_TSC2_Grabole-2016 1.9878918 1.39658395 2.860079 6.169167e-05
04_TSC2-tuber-Martin2017 3.5223279 0.93001955 9.482590 3.131705e-02
05_TSC2-SEN_SEGA-Martin2017 3.9779291 2.79667249 5.619997 5.063714e-14
06_all_EpilepsyGene 1.2436682 0.39536608 2.997206 6.067161e-01
07_high_conf_EpilepsyGene 2.3177651 0.46595788 7.073113 1.476893e-01
08_ASD_Epilepsy_EpilepsyGene 3.3640623 0.88889184 9.045974 3.596164e-02
09_Epi25_dataset 0.0000000 0.00000000 4.174176 1.000000e+00
10_SFARI_all 3.2829854 1.37375335 6.757351 4.526248e-03
11_SFARI_score4plus 3.8752839 1.50753939 8.368227 3.236208e-03
12_DeRubeis_RDNV 0.0000000 0.00000000 4.388931 1.000000e+00
13_Iossifov_RDNV 0.9063491 0.28856711 2.179380 1.000000e+00
14_FMRP_Darnell 2.0928373 1.15583616 3.550296 9.859691e-03
15_Voineagu_M12 0.8090892 0.16407906 2.432547 1.000000e+00
16_Voineagu_M16 1.3925198 0.44240092 3.359152 4.172680e-01
17_ID_genes_Neel 0.5699163 0.06807157 2.113266 5.887929e-01
18_ID_genes_pinto2014 0.7801407 0.09304023 2.900996 1.000000e+00
19_Fromer_SCZ_FunDN 1.5643803 0.65844116 3.194222 2.554450e-01
20_Cocchi_Pruning 5.5901824 1.74345774 13.871443 2.783365e-03
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.45175680 0.3370107 1.000000e+00
02_TSC2_KO_Grabole_2016 0.77555561 0.1729514 8.648660e-05
03_TSC2_Grabole-2016 0.68707467 0.1801252 1.233833e-03
04_TSC2-tuber-Martin2017 1.25912209 0.6794244 6.263410e-01
05_TSC2-SEN_SEGA-Martin2017 1.38076137 0.1797607 1.012743e-12
06_all_EpilepsyGene 0.21806524 0.5846982 1.000000e+00
07_high_conf_EpilepsyGene 0.84060339 0.8185018 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene 1.21314925 0.6790454 7.192327e-01
09_Epi25_dataset -Inf NaN 1.000000e+00
10_SFARI_all 1.18875318 0.4444931 9.052497e-02
11_SFARI_score4plus 1.35461893 0.4817042 6.472416e-02
12_DeRubeis_RDNV -Inf NaN 1.000000e+00
13_Iossifov_RDNV -0.09833069 0.5839270 1.000000e+00
14_FMRP_Darnell 0.73852070 0.3029065 1.971938e-01
15_Voineagu_M12 -0.21184616 0.8140616 1.000000e+00
16_Voineagu_M16 0.33111490 0.5850274 1.000000e+00
17_ID_genes_Neel -0.56226582 1.0841479 1.000000e+00
18_ID_genes_pinto2014 -0.24828105 1.0849195 1.000000e+00
19_Fromer_SCZ_FunDN 0.44748979 0.4415153 1.000000e+00
20_Cocchi_Pruning 1.72101191 0.5944600 5.566729e-02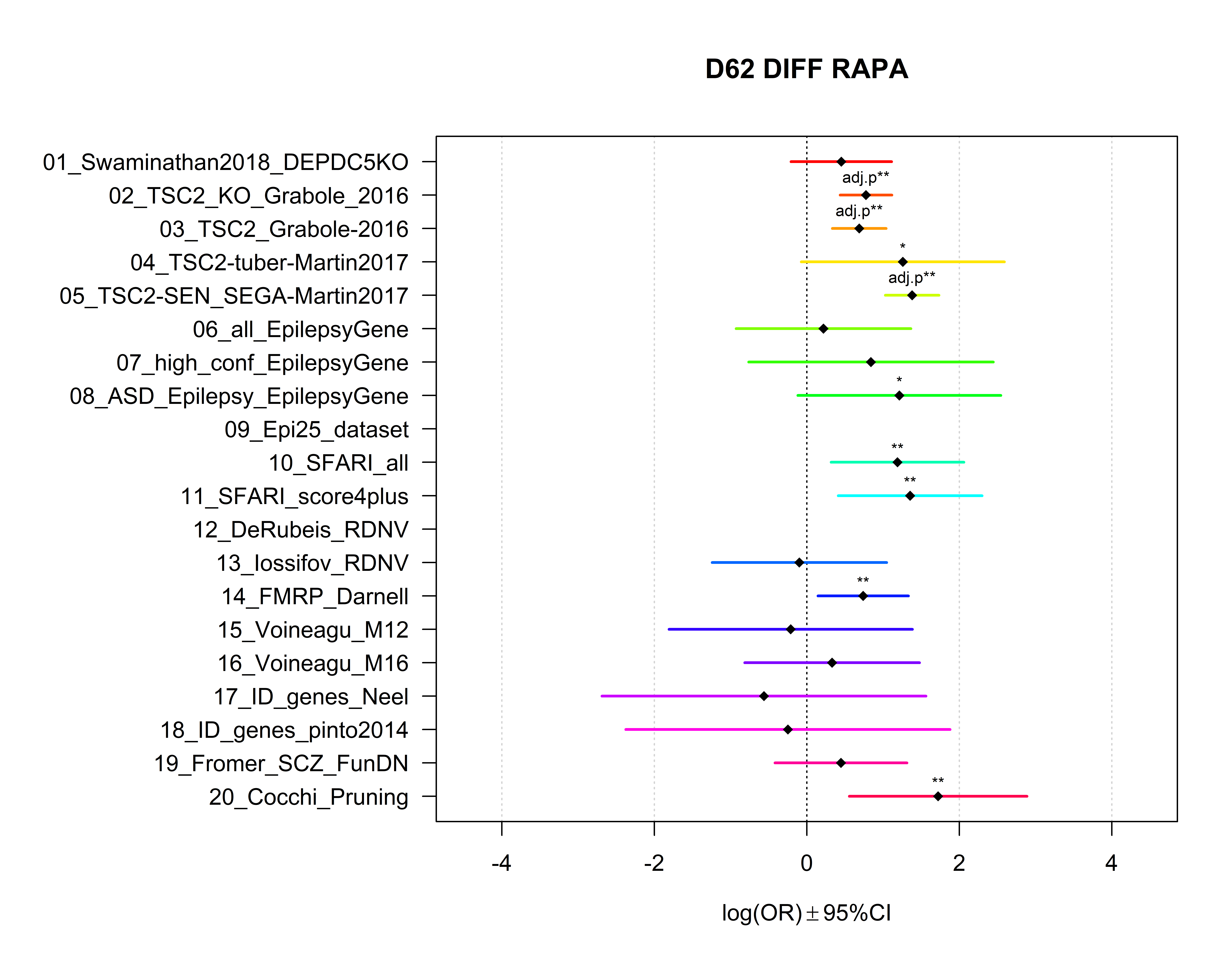
# with noRAPA NTCF as reference
noDIFFresults=data.frame(ENTREZ_ID=rownames(restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA),
SYMBOL=rowData(ddsMat)$hgnc,
REPLICATED = restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA$log2FoldChange*
restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_noRAPA$log2FoldChange>0 &
restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA$padj<=.05 &
restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_noRAPA$padj<=.05,
sg2.1_noDIFF_noRAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA$log2FoldChange,
sg2.1_noDIFF_noRAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA$padj,
sg2.2_noDIFF_noRAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_noRAPA$log2FoldChange,
sg2.2_noDIFF_noRAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_noRAPA$padj,
sgAVG_noDIFF_noRAPA_FC = rowMeans(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA$log2FoldChange,
restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_noRAPA$log2FoldChange)),
sgMax_noDIFF_noRAPA_adjP = rowMaxs(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_noRAPA$padj,
restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_noRAPA$padj)),
sg2.1_noDIFF_RAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_RAPA$log2FoldChange,
sg2.1_noDIFF_RAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_RAPA$padj,
sg2.2_noDIFF_RAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_RAPA$log2FoldChange,
sg2.2_noDIFF_RAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_RAPA$padj,
sgAVG_noDIFF_RAPA_FC = rowMeans(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_RAPA$log2FoldChange,
restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_RAPA$log2FoldChange)),
sgMax_noDIFF_RAPA_adjP = rowMaxs(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_noDIFF_RAPA$padj,
restab_D62_NTCnoRAPA_sgNTC_sg2.2_noDIFF_RAPA$padj)),
sg2.1_noDIFF_RAPA_rapantc_FC=restab_D62_NTCwRAPA_sgNTC_sg2.1_noDIFF_RAPA$log2FoldChange,
sg2.1_noDIFF_RAPA_rapantc_adjP=restab_D62_NTCwRAPA_sgNTC_sg2.1_noDIFF_RAPA$padj,
sg2.2_noDIFF_RAPA_rapantc_FC=restab_D62_NTCwRAPA_sgNTC_sg2.2_noDIFF_RAPA$log2FoldChange,
sg2.2_noDIFF_RAPA_rapantc_adjP=restab_D62_NTCwRAPA_sgNTC_sg2.2_noDIFF_RAPA$padj,
sgAVG_noDIFF_RAPA_rapantc_FC = rowMeans(cbind(restab_D62_NTCwRAPA_sgNTC_sg2.1_noDIFF_RAPA$log2FoldChange,
restab_D62_NTCwRAPA_sgNTC_sg2.2_noDIFF_RAPA$log2FoldChange)),
sgMax_noDIFF_RAPA_rapantc_adjP = rowMaxs(cbind(restab_D62_NTCwRAPA_sgNTC_sg2.1_noDIFF_RAPA$padj,
restab_D62_NTCwRAPA_sgNTC_sg2.2_noDIFF_RAPA$padj)))
for(i in names(genelists)){
noDIFFresults[,i] <- noDIFFresults$ENTREZ_ID %in% genelists[[i]]$ENTREZ_ID
}
write.xlsx(noDIFFresults, file=paste0(output, "/noDIFF_refnoRAPA_allgenes.xlsx"))
lm(noDIFFresults$sgAVG_noDIFF_noRAPA_FC~noDIFFresults$sgAVG_noDIFF_RAPA_FC) %>% summary()
Call:
lm(formula = noDIFFresults$sgAVG_noDIFF_noRAPA_FC ~ noDIFFresults$sgAVG_noDIFF_RAPA_FC)
Residuals:
Min 1Q Median 3Q Max
-6.6160 -0.2293 0.0369 0.2866 4.8184
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.032691 0.005973 -5.473 4.5e-08 ***
noDIFFresults$sgAVG_noDIFF_RAPA_FC 0.533189 0.006943 76.798 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.6987 on 13707 degrees of freedom
(22 Beobachtungen als fehlend gelöscht)
Multiple R-squared: 0.3008, Adjusted R-squared: 0.3008
F-statistic: 5898 on 1 and 13707 DF, p-value: < 2.2e-16DIFFresults=data.frame(ENTREZ_ID=rownames(restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA),
SYMBOL=rowData(ddsMat)$hgnc,
REPLICATED = restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA$log2FoldChange*
restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_noRAPA$log2FoldChange>0 &
restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA$padj<=.05 &
restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_noRAPA$padj<=.05,
sg2.1_DIFF_noRAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA$log2FoldChange,
sg2.1_DIFF_noRAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA$padj,
sg2.2_DIFF_noRAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_noRAPA$log2FoldChange,
sg2.2_DIFF_noRAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_noRAPA$padj,
sgAVG_DIFF_noRAPA_FC = rowMeans(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA$log2FoldChange, restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_noRAPA$log2FoldChange)),
sgMAX_DIFF_noRAPA_adjP = rowMaxs(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_noRAPA$padj, restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_noRAPA$padj)),
sg2.1_DIFF_RAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_RAPA$log2FoldChange,
sg2.1_DIFF_RAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_RAPA$padj,
sg2.2_DIFF_RAPA_FC=restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_RAPA$log2FoldChange,
sg2.2_DIFF_RAPA_adjP=restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_RAPA$padj,
sgAVG_DIFF_RAPA_FC = rowMeans(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_RAPA$log2FoldChange, restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_RAPA$log2FoldChange)),
sgMAX_DIFF_RAPA_adjP = rowMaxs(cbind(restab_D62_NTCnoRAPA_sgNTC_sg2.1_DIFF_RAPA$padj, restab_D62_NTCnoRAPA_sgNTC_sg2.2_DIFF_RAPA$padj)),
sg2.1_DIFF_RAPA_rapantc_FC=restab_D62_NTCwRAPA_sgNTC_sg2.1_DIFF_RAPA$log2FoldChange,
sg2.1_DIFF_RAPA_rapantc_adjP=restab_D62_NTCwRAPA_sgNTC_sg2.1_DIFF_RAPA$padj,
sg2.2_DIFF_RAPA_rapantc_FC=restab_D62_NTCwRAPA_sgNTC_sg2.2_DIFF_RAPA$log2FoldChange,
sg2.2_DIFF_RAPA_rapantc_adjP=restab_D62_NTCwRAPA_sgNTC_sg2.2_DIFF_RAPA$padj,
sgAVG_DIFF_RAPA_rapantc_FC = rowMeans(cbind(restab_D62_NTCwRAPA_sgNTC_sg2.1_DIFF_RAPA$log2FoldChange, restab_D62_NTCwRAPA_sgNTC_sg2.2_DIFF_RAPA$log2FoldChange)),
sgMAX_DIFF_RAPA_rapantc_adjP = rowMaxs(cbind(restab_D62_NTCwRAPA_sgNTC_sg2.1_DIFF_RAPA$padj, restab_D62_NTCwRAPA_sgNTC_sg2.2_DIFF_RAPA$padj)))
for(i in names(genelists)){
DIFFresults[,i] <- DIFFresults$ENTREZ_ID %in% genelists[[i]]$ENTREZ_ID
}
metadata <- mcols(ddsMat) %>% as.data.frame()
for(i in names(genelists)){
metadata[,i] <- rownames(metadata) %in% genelists[[i]]$ENTREZ_ID
}
mcols(ddsMat) <- metadata
write.xlsx(DIFFresults, file=paste0(output, "/DIFF_refnoRAPA_allgenes.xlsx"))
# check if the same genes are differentially reguatled
lm(DIFFresults$sgAVG_DIFF_noRAPA_FC~DIFFresults$sgAVG_DIFF_RAPA_FC) %>% summary()
Call:
lm(formula = DIFFresults$sgAVG_DIFF_noRAPA_FC ~ DIFFresults$sgAVG_DIFF_RAPA_FC)
Residuals:
Min 1Q Median 3Q Max
-9.9544 -0.2712 -0.0270 0.2281 5.7756
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.006635 0.005412 1.226 0.22
DIFFresults$sgAVG_DIFF_RAPA_FC 0.728655 0.005332 136.664 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.6322 on 13705 degrees of freedom
(24 Beobachtungen als fehlend gelöscht)
Multiple R-squared: 0.5768, Adjusted R-squared: 0.5767
F-statistic: 1.868e+04 on 1 and 13705 DF, p-value: < 2.2e-16noDIFFhits=(noDIFFresults$sg2.1_noDIFF_noRAPA_adjP<=0.05 &
noDIFFresults$sg2.2_noDIFF_noRAPA_adjP<=0.05 &
sign(noDIFFresults$sg2.1_noDIFF_noRAPA_FC)==sign(noDIFFresults$sg2.2_noDIFF_noRAPA_FC))
sum(noDIFFhits, na.rm = T)[1] 237sum(noDIFFhits & noDIFFresults$sg2.1_noDIFF_noRAPA_FC>0, na.rm = T)[1] 139sum(noDIFFhits & noDIFFresults$sg2.1_noDIFF_noRAPA_FC<0, na.rm = T)[1] 98ggplot(data = noDIFFresults[,], aes(sg2.1_noDIFF_noRAPA_FC, sg2.2_noDIFF_noRAPA_FC, col=noDIFFhits)) +
geom_point(pch=16, alpha=0.5) +
geom_abline(slope=1, intercept = 0) + ggtitle("DIFF correlation between KOs")Warning: Removed 15 rows containing missing values (`geom_point()`).
## rescued Genes
noDIFF_RAPA_nosig = (noDIFFresults$sg2.1_noDIFF_RAPA_adjP>0.05 & noDIFFresults$sg2.2_noDIFF_RAPA_adjP>0.05)
# just necessary to check one KO as significant DEX is defoned that both have to have the same direction
noDIFF_RAPA_chngsFC = (sign(noDIFFresults$sg2.1_noDIFF_noRAPA_FC) != sign(noDIFFresults$sg2.1_noDIFF_RAPA_FC)) &
(sign(noDIFFresults$sg2.1_noDIFF_noRAPA_FC) != sign(noDIFFresults$sg2.2_noDIFF_RAPA_FC))
noDIFFhits_ENTREZ = na.omit(noDIFFresults$ENTREZ_ID[noDIFFhits])
noDIFFRAPA_RESC_ENTREZ = na.omit(noDIFFresults$ENTREZ_ID[ (noDIFFhits & noDIFF_RAPA_nosig)| (noDIFFhits & noDIFF_RAPA_chngsFC)])
noDIFFRAPA_noRESC_ENTREZ = noDIFFhits_ENTREZ[! noDIFFhits_ENTREZ %in% noDIFFRAPA_RESC_ENTREZ]
GOIs =list(noDIFFRAPA_RESC=noDIFFRAPA_RESC_ENTREZ,
noDIFFRAPA_noRESC=noDIFFRAPA_noRESC_ENTREZ)
Rescue_Genelist = list(PROLIF=GOIs)
for (n in names(GOIs)){
Resall=data.frame()
for (gl in names(genelists)){
tmp=table(signif=geneids %in% GOIs[[n]], targetgene=geneids %in% genelists[[gl]]$ENTREZ_ID)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resall = rbind(Resall, res)
}
colnames(Resall)=c("OR", "CI95L", "CI95U", "P")
rownames(Resall)=names(genelists)
Resall$Beta = log(Resall$OR)
Resall$SE = (log(Resall$OR)-log(Resall$CI95L))/1.96
Resall$Padj=p.adjust(Resall$P, method = "bonferroni")
print(Resall)
multiORplot(Resall, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE",
pheno=n, xlims=c(-4.5,4.5))
} OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.8962220 0.911530751 3.568958 0.056083599
02_TSC2_KO_Grabole_2016 1.2217549 0.803375513 1.839685 0.353405161
03_TSC2_Grabole-2016 1.2100031 0.808429778 1.817862 0.378031656
04_TSC2-tuber-Martin2017 5.0017523 1.313509365 13.581994 0.010350766
05_TSC2-SEN_SEGA-Martin2017 1.9776818 1.214996272 3.124058 0.004642208
06_all_EpilepsyGene 0.3370130 0.008432705 1.931077 0.374198551
07_high_conf_EpilepsyGene 0.0000000 0.000000000 3.951174 1.000000000
08_ASD_Epilepsy_EpilepsyGene 1.1296704 0.028090902 6.560011 0.591072462
09_Epi25_dataset 1.5688392 0.038885691 9.178556 0.476267938
10_SFARI_all 1.6473395 0.331853744 5.011086 0.435882613
11_SFARI_score4plus 0.7223500 0.018016559 4.166233 1.000000000
12_DeRubeis_RDNV 1.6487664 0.040842341 9.659725 0.459837758
13_Iossifov_RDNV 1.5681418 0.559278472 3.560051 0.291371543
14_FMRP_Darnell 1.4020081 0.586416629 2.888490 0.386785201
15_Voineagu_M12 0.3723823 0.009314882 2.135126 0.525734045
16_Voineagu_M16 2.8509610 1.108347992 6.163063 0.015440752
17_ID_genes_Neel 0.3972944 0.009935966 2.278697 0.522903902
18_ID_genes_pinto2014 0.5432259 0.013569641 3.123350 1.000000000
19_Fromer_SCZ_FunDN 1.9435398 0.757336454 4.188107 0.103209798
20_Cocchi_Pruning 2.9867194 0.351472776 11.388948 0.150938725
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.6398635 0.3737211 1.00000000
02_TSC2_KO_Grabole_2016 0.2002883 0.2138884 1.00000000
03_TSC2_Grabole-2016 0.1906229 0.2057573 1.00000000
04_TSC2-tuber-Martin2017 1.6097883 0.6821867 0.20701531
05_TSC2-SEN_SEGA-Martin2017 0.6819253 0.2485634 0.09284417
06_all_EpilepsyGene -1.0876338 1.8816347 1.00000000
07_high_conf_EpilepsyGene -Inf NaN 1.00000000
08_ASD_Epilepsy_EpilepsyGene 0.1219259 1.8848140 1.00000000
09_Epi25_dataset 0.4503360 1.8864617 1.00000000
10_SFARI_all 0.4991616 0.8174605 1.00000000
11_SFARI_score4plus -0.3252454 1.8832748 1.00000000
12_DeRubeis_RDNV 0.5000274 1.8867670 1.00000000
13_Iossifov_RDNV 0.4498913 0.5260199 1.00000000
14_FMRP_Darnell 0.3379056 0.4447094 1.00000000
15_Voineagu_M12 -0.9878344 1.8817896 1.00000000
16_Voineagu_M16 1.0476561 0.4820334 0.30881503
17_ID_genes_Neel -0.9230777 1.8818962 1.00000000
18_ID_genes_pinto2014 -0.6102301 1.8824950 1.00000000
19_Fromer_SCZ_FunDN 0.6645109 0.4808462 1.00000000
20_Cocchi_Pruning 1.0941756 1.0917340 1.00000000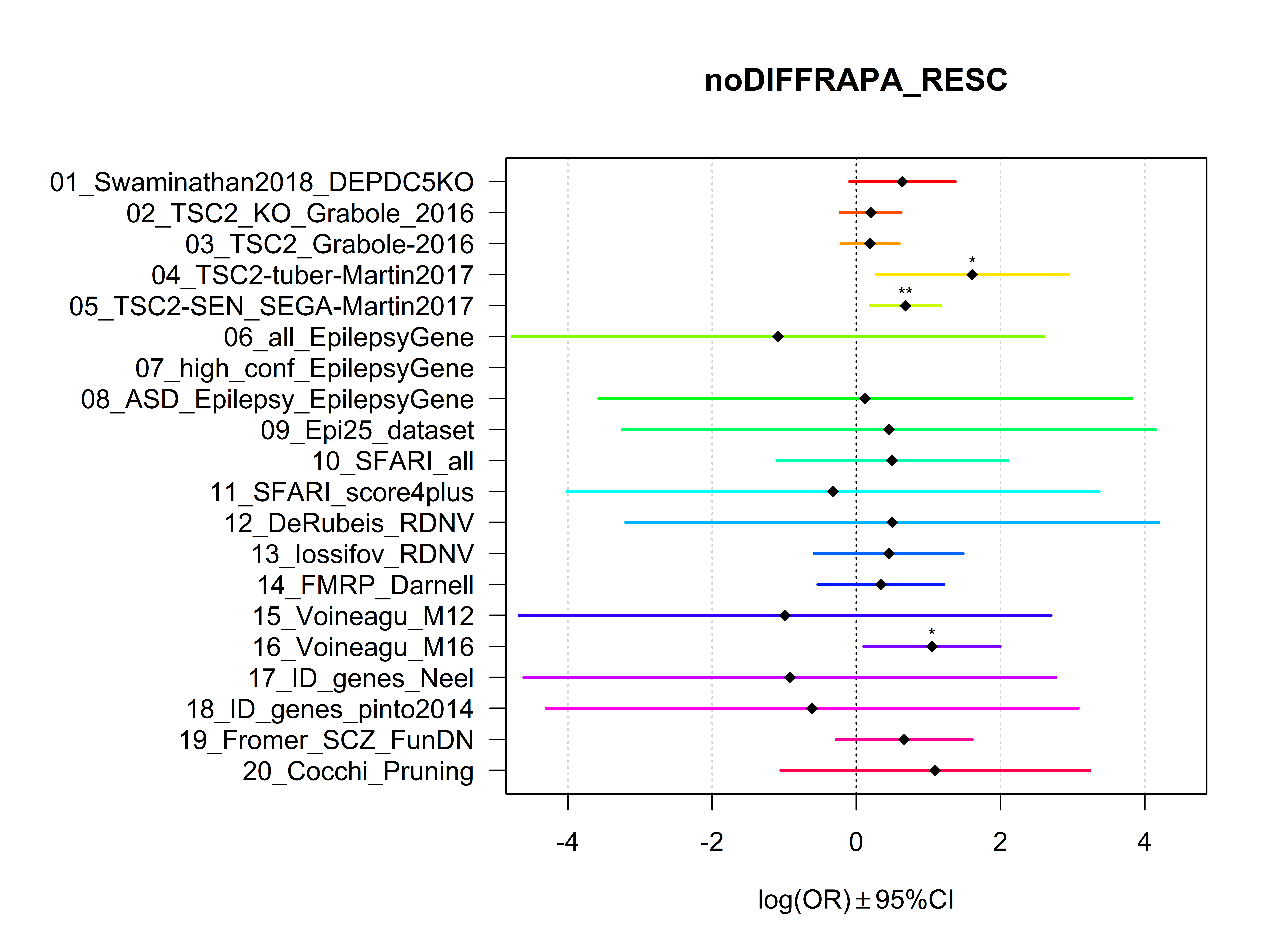
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 0.7648963 0.27463081 1.719572 7.078505e-01
02_TSC2_KO_Grabole_2016 2.3047717 1.61017655 3.309040 2.063852e-06
03_TSC2_Grabole-2016 2.8460661 1.91234218 4.323870 2.153124e-08
04_TSC2-tuber-Martin2017 2.9045975 0.58222672 8.907576 9.140780e-02
05_TSC2-SEN_SEGA-Martin2017 4.7901632 3.32477250 6.873501 1.507645e-16
06_all_EpilepsyGene 1.3921050 0.44178111 3.363956 4.173341e-01
07_high_conf_EpilepsyGene 1.6994039 0.20136399 6.395293 3.351302e-01
08_ASD_Epilepsy_EpilepsyGene 2.7750826 0.55664935 8.501253 1.010018e-01
09_Epi25_dataset 0.0000000 0.00000000 4.660279 1.000000e+00
10_SFARI_all 1.2999599 0.26275219 3.931033 5.066738e-01
11_SFARI_score4plus 1.7634015 0.35556928 5.354192 2.499747e-01
12_DeRubeis_RDNV 0.0000000 0.00000000 4.900161 1.000000e+00
13_Iossifov_RDNV 2.1338933 0.99201771 4.092438 3.257362e-02
14_FMRP_Darnell 1.5486293 0.74975566 2.886936 1.773306e-01
15_Voineagu_M12 0.5963721 0.07117314 2.214779 7.758121e-01
16_Voineagu_M16 2.9325996 1.29844787 5.818451 5.555544e-03
17_ID_genes_Neel 1.3009101 0.34699517 3.445838 5.557777e-01
18_ID_genes_pinto2014 0.4303979 0.01077268 2.463698 7.313735e-01
19_Fromer_SCZ_FunDN 1.0645640 0.33823760 2.567505 8.122055e-01
20_Cocchi_Pruning 3.6115255 0.72120512 11.144081 5.579388e-02
Beta SE Padj
01_Swaminathan2018_DEPDC5KO -0.26801505 0.5226084 1.000000e+00
02_TSC2_KO_Grabole_2016 0.83498163 0.1829785 4.127704e-05
03_TSC2_Grabole-2016 1.04593772 0.2028617 4.306248e-07
04_TSC2-tuber-Martin2017 1.06629483 0.8199950 1.000000e+00
05_TSC2-SEN_SEGA-Martin2017 1.56656448 0.1863078 3.015290e-15
06_all_EpilepsyGene 0.33081701 0.5855907 1.000000e+00
07_high_conf_EpilepsyGene 0.53027755 1.0882238 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene 1.02068053 0.8196430 1.000000e+00
09_Epi25_dataset -Inf NaN 1.000000e+00
10_SFARI_all 0.26233340 0.8157537 1.000000e+00
11_SFARI_score4plus 0.56724464 0.8169795 1.000000e+00
12_DeRubeis_RDNV -Inf NaN 1.000000e+00
13_Iossifov_RDNV 0.75794817 0.3907972 6.514723e-01
14_FMRP_Darnell 0.43737020 0.3700909 1.000000e+00
15_Voineagu_M12 -0.51689051 1.0845659 1.000000e+00
16_Voineagu_M16 1.07588925 0.4156733 1.111109e-01
17_ID_genes_Neel 0.26306409 0.6742390 1.000000e+00
18_ID_genes_pinto2014 -0.84304520 1.8814781 1.000000e+00
19_Fromer_SCZ_FunDN 0.06256531 0.5849857 1.000000e+00
20_Cocchi_Pruning 1.28413025 0.8219194 1.000000e+00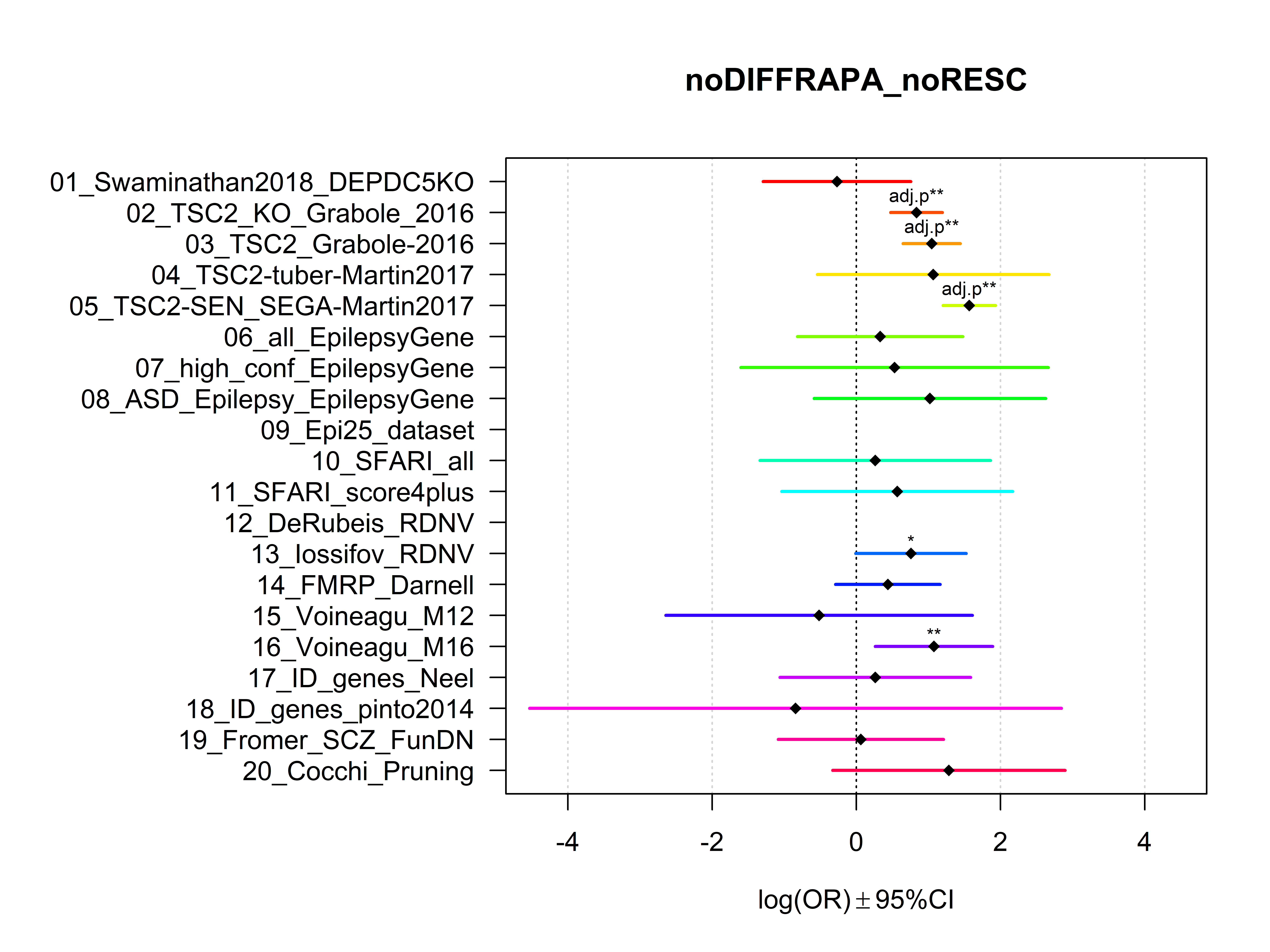
gene_univers = rownames(ddsMat)
gostres = getGOresults(GOIs, gene_univers)
toptab = gostres$result
write.xlsx(toptab, file = paste0(output, "/GOres_Rescue_noDIFF.xlsx"), sheetName = "GO_enrichment")
fit<- euler(list(PROLIF_DEX=noDIFFhits_ENTREZ,
RAPA_RESC=noDIFFRAPA_RESC_ENTREZ), shape = "ellipse")
plot(fit, quantities = TRUE, main="PROLIF rescue")
### DIFFERERNTIATION
DIFFhits=(DIFFresults$sg2.1_DIFF_noRAPA_adjP<=0.05 &
DIFFresults$sg2.2_DIFF_noRAPA_adjP<=0.05 &
sign(DIFFresults$sg2.1_DIFF_noRAPA_FC)==sign(DIFFresults$sg2.2_DIFF_noRAPA_FC))
sum(DIFFhits, na.rm = T)[1] 570sum(DIFFhits & DIFFresults$sg2.1_DIFF_noRAPA_FC>0, na.rm = T)[1] 250sum(DIFFhits & DIFFresults$sg2.1_DIFF_noRAPA_FC<0, na.rm = T)[1] 320ggplot(data = DIFFresults[,], aes(sg2.1_DIFF_noRAPA_FC, sg2.2_DIFF_noRAPA_FC, col=DIFFhits)) +
geom_point(pch=16, alpha=0.5) +
geom_abline(slope=1, intercept = 0) +
theme_bw()Warning: Removed 9 rows containing missing values (`geom_point()`).
## rescued Genes
DIFF_RAPA_nosig = (DIFFresults$sg2.1_DIFF_RAPA_adjP>0.05 & DIFFresults$sg2.2_DIFF_RAPA_adjP>0.05)
# just necessary to check one KO as significant DEX is defoned that both have to have the same direction
DIFF_RAPA_chngsFC = (sign(DIFFresults$sg2.1_DIFF_noRAPA_FC) != sign(DIFFresults$sg2.1_DIFF_RAPA_FC)) &
(sign(DIFFresults$sg2.1_DIFF_noRAPA_FC) != sign(DIFFresults$sg2.2_DIFF_RAPA_FC))
DIFFhits_ENTREZ = na.omit(DIFFresults$ENTREZ_ID[DIFFhits])
DIFFRAPA_RESC_ENTREZ = na.omit(DIFFresults$ENTREZ_ID[ (DIFFhits & DIFF_RAPA_nosig)| (DIFFhits & DIFF_RAPA_chngsFC)])
DIFFRAPA_noRESC_ENTREZ = DIFFhits_ENTREZ[! DIFFhits_ENTREZ %in% DIFFRAPA_RESC_ENTREZ]
GOIs =list(DIFFRAPA_RESC=DIFFRAPA_RESC_ENTREZ,
DIFFRAPA_noRESC=DIFFRAPA_noRESC_ENTREZ)
Rescue_Genelist[["DIFF"]] = GOIs
for (n in names(GOIs)){
Resall=data.frame()
for (gl in names(genelists)){
tmp=table(signif=geneids %in% GOIs[[n]], targetgene=geneids %in% genelists[[gl]]$ENTREZ_ID)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resall = rbind(Resall, res)
}
colnames(Resall)=c("OR", "CI95L", "CI95U", "P")
rownames(Resall)=names(genelists)
Resall$Beta = log(Resall$OR)
Resall$SE = (log(Resall$OR)-log(Resall$CI95L))/1.96
Resall$Padj=p.adjust(Resall$P, method = "bonferroni")
print(Resall)
multiORplot(Resall, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE",
pheno=n, xlims=c(-4.5,4.5))
} OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.8891393 1.19980453 2.863444 4.104511e-03
02_TSC2_KO_Grabole_2016 1.9445559 1.50211707 2.517220 2.735000e-07
03_TSC2_Grabole-2016 1.8078886 1.38579985 2.370461 5.494935e-06
04_TSC2-tuber-Martin2017 2.5641410 0.80864668 6.255024 5.288820e-02
05_TSC2-SEN_SEGA-Martin2017 2.7611616 2.07634220 3.644987 8.353515e-12
06_all_EpilepsyGene 0.7122637 0.22787876 1.699957 5.624527e-01
07_high_conf_EpilepsyGene 0.4343922 0.01086968 2.488120 7.305864e-01
08_ASD_Epilepsy_EpilepsyGene 0.0000000 0.00000000 1.733132 2.795309e-01
09_Epi25_dataset 0.0000000 0.00000000 2.413608 4.099390e-01
10_SFARI_all 1.8635289 0.78616580 3.791970 8.781811e-02
11_SFARI_score4plus 2.2065886 0.86491367 4.713672 4.753787e-02
12_DeRubeis_RDNV 2.1054608 0.42202664 6.458896 1.801229e-01
13_Iossifov_RDNV 1.3006993 0.65818798 2.335348 3.971279e-01
14_FMRP_Darnell 1.4799304 0.88203383 2.356335 9.571302e-02
15_Voineagu_M12 0.9522070 0.34359677 2.124533 1.000000e+00
16_Voineagu_M16 3.9755890 2.40891783 6.271949 2.902900e-07
17_ID_genes_Neel 1.1950946 0.47154511 2.529327 5.322089e-01
18_ID_genes_pinto2014 1.1555563 0.36842197 2.772631 6.264708e-01
19_Fromer_SCZ_FunDN 1.0050600 0.45126753 1.956236 1.000000e+00
20_Cocchi_Pruning 1.8621430 0.37415993 5.688552 2.271892e-01
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.636121330 0.2316136 8.209022e-02
02_TSC2_KO_Grabole_2016 0.665033625 0.1317133 5.470001e-06
03_TSC2_Grabole-2016 0.592159642 0.1356542 1.098987e-04
04_TSC2-tuber-Martin2017 0.941623513 0.5887840 1.000000e+00
05_TSC2-SEN_SEGA-Martin2017 1.015651450 0.1454304 1.670703e-10
06_all_EpilepsyGene -0.339307129 0.5814461 1.000000e+00
07_high_conf_EpilepsyGene -0.833807567 1.8816175 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene -Inf NaN 1.000000e+00
09_Epi25_dataset -Inf NaN 1.000000e+00
10_SFARI_all 0.622471973 0.4403365 1.000000e+00
11_SFARI_score4plus 0.791447723 0.4778435 9.507575e-01
12_DeRubeis_RDNV 0.744534364 0.8200108 1.000000e+00
13_Iossifov_RDNV 0.262902039 0.3475341 1.000000e+00
14_FMRP_Darnell 0.391995032 0.2640408 1.000000e+00
15_Voineagu_M12 -0.048972791 0.5200580 1.000000e+00
16_Voineagu_M16 1.380172907 0.2556098 5.805801e-06
17_ID_genes_Neel 0.178225323 0.4744724 1.000000e+00
18_ID_genes_pinto2014 0.144581894 0.5832185 1.000000e+00
19_Fromer_SCZ_FunDN 0.005047199 0.4085419 1.000000e+00
20_Cocchi_Pruning 0.621727962 0.8187755 1.000000e+00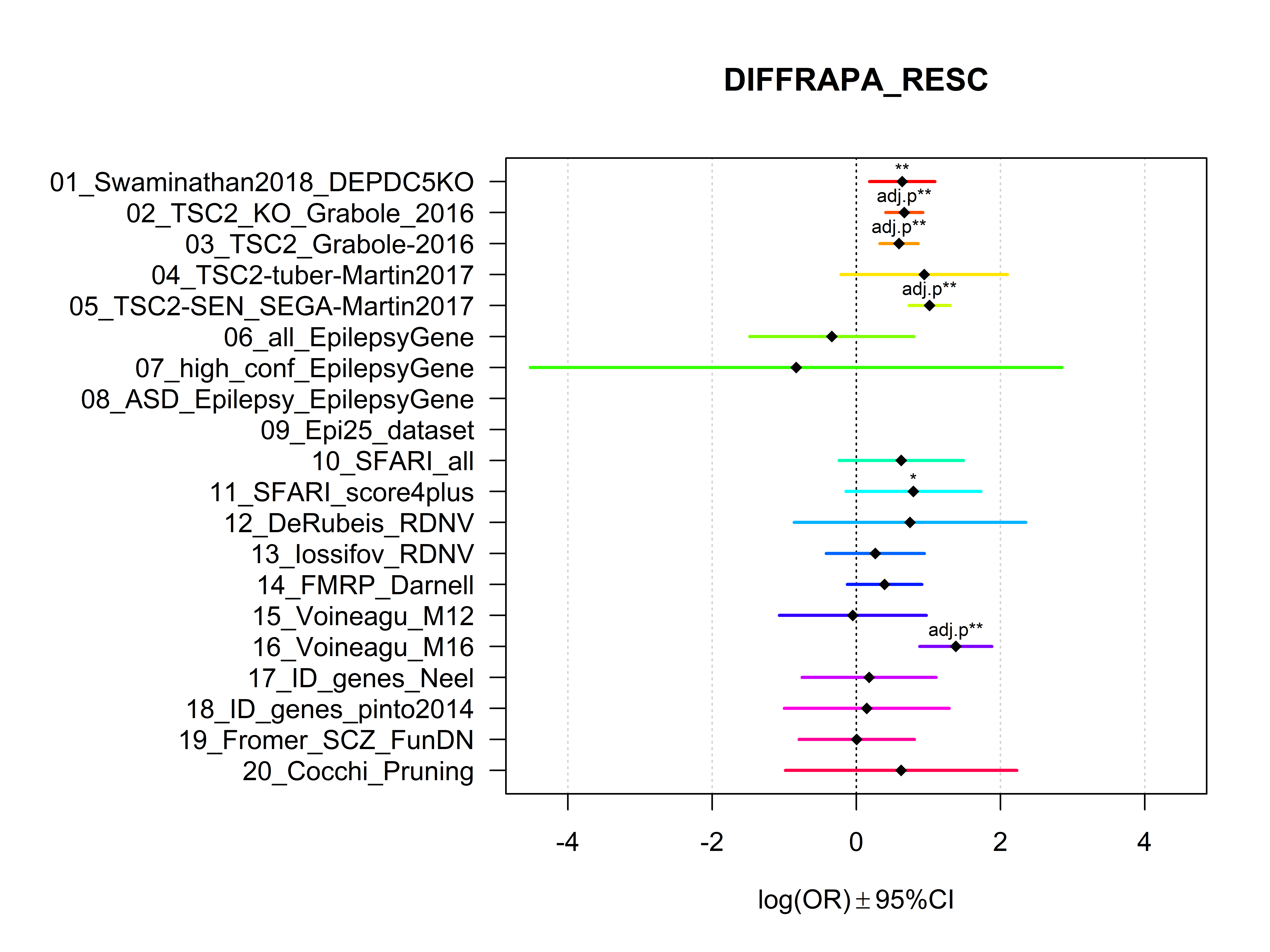
OR CI95L CI95U P
01_Swaminathan2018_DEPDC5KO 1.3808266 0.8734614 2.095386 1.454025e-01
02_TSC2_KO_Grabole_2016 1.6907262 1.3435532 2.125508 5.263292e-06
03_TSC2_Grabole-2016 1.9351144 1.5256057 2.465333 1.587112e-08
04_TSC2-tuber-Martin2017 4.2640338 1.9653149 8.273783 2.576900e-04
05_TSC2-SEN_SEGA-Martin2017 2.8180606 2.1903830 3.605470 4.686266e-15
06_all_EpilepsyGene 1.3893244 0.7034997 2.492215 2.948316e-01
07_high_conf_EpilepsyGene 2.1594725 0.7708589 4.895185 6.917868e-02
08_ASD_Epilepsy_EpilepsyGene 1.5078515 0.4013453 4.007951 3.483485e-01
09_Epi25_dataset 1.0256268 0.1216163 3.855898 7.225946e-01
10_SFARI_all 0.8852094 0.2828034 2.117279 1.000000e+00
11_SFARI_score4plus 0.7098934 0.1442467 2.127879 8.020013e-01
12_DeRubeis_RDNV 2.2281418 0.5882687 5.999134 1.154765e-01
13_Iossifov_RDNV 1.2794987 0.7019030 2.165088 3.671954e-01
14_FMRP_Darnell 1.3267725 0.8226961 2.045149 2.146901e-01
15_Voineagu_M12 1.1332862 0.5088418 2.205854 7.146470e-01
16_Voineagu_M16 2.5825756 1.5131714 4.170728 3.850128e-04
17_ID_genes_Neel 1.2110133 0.5434303 2.358910 5.714451e-01
18_ID_genes_pinto2014 0.9006786 0.2877019 2.154842 1.000000e+00
19_Fromer_SCZ_FunDN 0.7804990 0.3513532 1.513744 6.449328e-01
20_Cocchi_Pruning 5.3911724 2.4663155 10.571777 4.392584e-05
Beta SE Padj
01_Swaminathan2018_DEPDC5KO 0.32268233 0.2336600 1.000000e+00
02_TSC2_KO_Grabole_2016 0.52515812 0.1172655 1.052658e-04
03_TSC2_Grabole-2016 0.66016643 0.1213137 3.174225e-07
04_TSC2-tuber-Martin2017 1.45021561 0.3951853 5.153800e-03
05_TSC2-SEN_SEGA-Martin2017 1.03604892 0.1285574 9.372532e-14
06_all_EpilepsyGene 0.32881755 0.3471966 1.000000e+00
07_high_conf_EpilepsyGene 0.76986397 0.5255683 1.000000e+00
08_ASD_Epilepsy_EpilepsyGene 0.41068582 0.6753157 1.000000e+00
09_Epi25_dataset 0.02530394 1.0878512 1.000000e+00
10_SFARI_all -0.12193103 0.5821798 1.000000e+00
11_SFARI_score4plus -0.34264041 0.8130560 1.000000e+00
12_DeRubeis_RDNV 0.80116795 0.6794589 1.000000e+00
13_Iossifov_RDNV 0.24646833 0.3063410 1.000000e+00
14_FMRP_Darnell 0.28274932 0.2438356 1.000000e+00
15_Voineagu_M12 0.12512153 0.4085406 1.000000e+00
16_Voineagu_M16 0.94878720 0.2727446 7.700256e-03
17_ID_genes_Neel 0.19145747 0.4088323 1.000000e+00
18_ID_genes_pinto2014 -0.10460677 0.5822570 1.000000e+00
19_Fromer_SCZ_FunDN -0.24782181 0.4072151 1.000000e+00
20_Cocchi_Pruning 1.68476288 0.3989987 8.785167e-04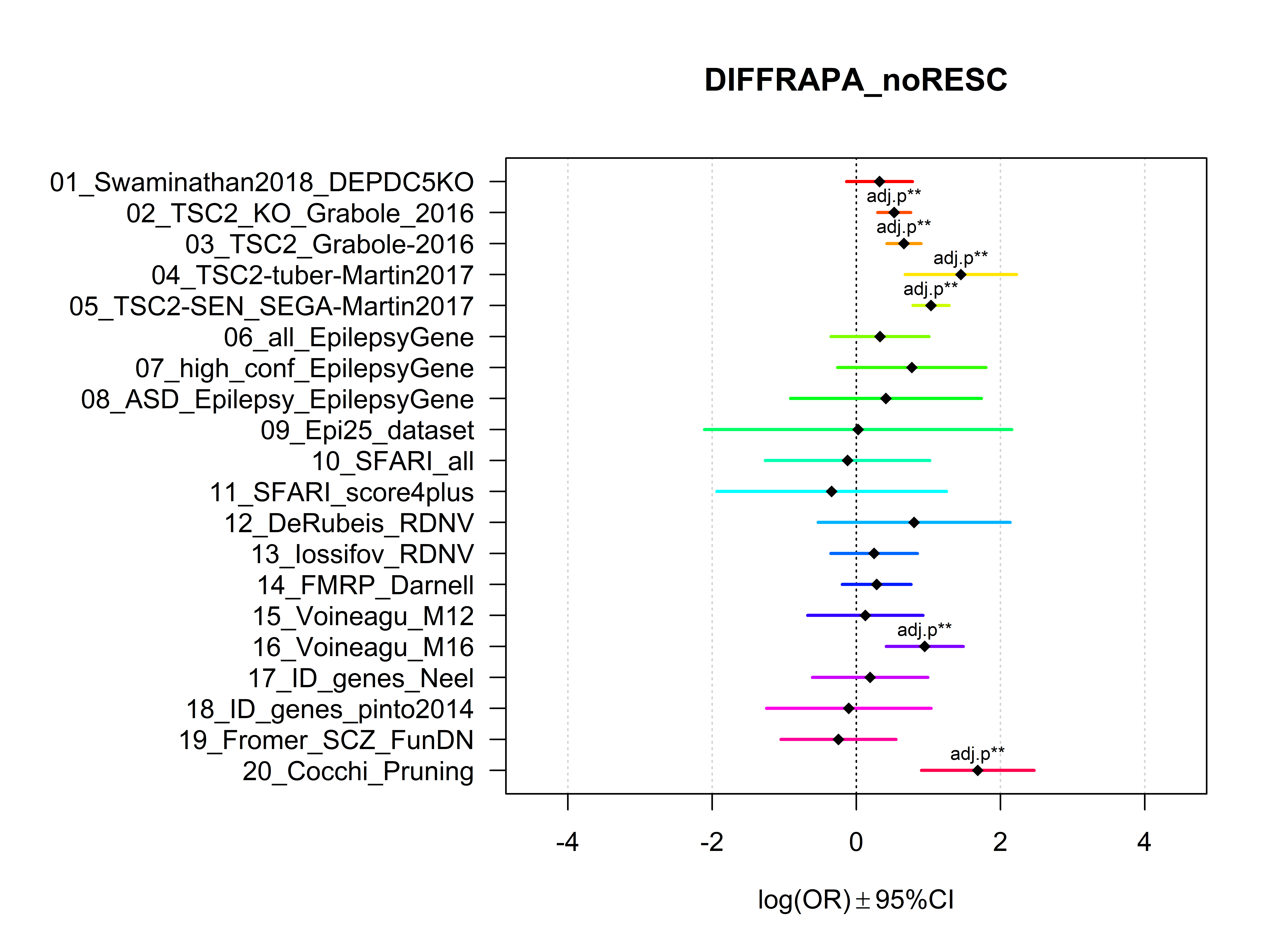
gene_univers = rownames(ddsMat)
gostres = getGOresults(GOIs, gene_univers)
toptab = gostres$result
write.xlsx(toptab, file = paste0(output, "/GOres_Rescue_DIFF.xlsx"), sheetName = "GO_enrichment")
fit<- euler(list(DIFF_DEX=DIFFhits_ENTREZ,
RAPA_RESC=DIFFRAPA_RESC_ENTREZ), shape = "ellipse")
plot(fit, quantities = TRUE, main="DIFF rescue")
allhits = list(DIFF_DEX=DIFFhits_ENTREZ,
DIFF_RAPA_RESC=DIFFRAPA_RESC_ENTREZ,
PROLIF_DEX=noDIFFhits_ENTREZ,
PROLIF_RAPA_RESC=noDIFFRAPA_RESC_ENTREZ
)
fit2 <- euler(allhits, shape = "ellipse")
plot(fit2, quantities = TRUE, main="Overlap of gene hits")
save(ddsMat, file="./output/D62_dds_matrix.RData")
save(Rescue_Genelist, file = "./output/RescueGenelists.RData")
sessionInfo()R version 4.2.0 (2022-04-22 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=German_Germany.utf8 LC_CTYPE=German_Germany.utf8
[3] LC_MONETARY=German_Germany.utf8 LC_NUMERIC=C
[5] LC_TIME=German_Germany.utf8
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] gprofiler2_0.2.1 eulerr_6.1.1
[3] openxlsx_4.2.5 limma_3.52.3
[5] flashClust_1.01-2 WGCNA_1.71
[7] fastcluster_1.2.3 dynamicTreeCut_1.63-1
[9] knitr_1.42 DESeq2_1.36.0
[11] SummarizedExperiment_1.26.1 Biobase_2.56.0
[13] MatrixGenerics_1.8.1 matrixStats_0.63.0
[15] GenomicRanges_1.48.0 GenomeInfoDb_1.32.4
[17] IRanges_2.30.1 S4Vectors_0.34.0
[19] BiocGenerics_0.42.0 pheatmap_1.0.12
[21] RColorBrewer_1.1-3 compareGroups_4.5.1
[23] forcats_1.0.0 stringr_1.5.0
[25] dplyr_1.1.0 purrr_1.0.1
[27] readr_2.1.3 tidyr_1.3.0
[29] tibble_3.1.8 ggplot2_3.4.0
[31] tidyverse_1.3.2 kableExtra_1.3.4
[33] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] utf8_1.2.2 tidyselect_1.2.0 RSQLite_2.2.20
[4] AnnotationDbi_1.58.0 htmlwidgets_1.6.1 grid_4.2.0
[7] BiocParallel_1.30.3 munsell_0.5.0 codetools_0.2-18
[10] preprocessCore_1.58.0 interp_1.1-3 chron_2.3-57
[13] withr_2.5.0 colorspace_2.1-0 highr_0.10
[16] uuid_1.1-0 rstudioapi_0.14 officer_0.4.4
[19] labeling_0.4.2 git2r_0.30.1 GenomeInfoDbData_1.2.8
[22] polyclip_1.10-0 farver_2.1.1 bit64_4.0.5
[25] rprojroot_2.0.3 vctrs_0.5.2 generics_0.1.3
[28] xfun_0.36 timechange_0.2.0 R6_2.5.1
[31] doParallel_1.0.17 locfit_1.5-9.6 bitops_1.0-7
[34] cachem_1.0.6 DelayedArray_0.22.0 assertthat_0.2.1
[37] promises_1.2.0.1 scales_1.2.1 nnet_7.3-17
[40] googlesheets4_1.0.1 gtable_0.3.1 processx_3.7.0
[43] rlang_1.0.6 genefilter_1.78.0 systemfonts_1.0.4
[46] splines_4.2.0 lazyeval_0.2.2 gargle_1.3.0
[49] impute_1.70.0 broom_1.0.3 checkmate_2.1.0
[52] yaml_2.3.7 modelr_0.1.10 backports_1.4.1
[55] httpuv_1.6.8 HardyWeinberg_1.7.5 Hmisc_4.7-1
[58] tools_4.2.0 ellipsis_0.3.2 jquerylib_0.1.4
[61] Rsolnp_1.16 Rcpp_1.0.10 base64enc_0.1-3
[64] zlibbioc_1.42.0 RCurl_1.98-1.8 ps_1.7.1
[67] rpart_4.1.16 deldir_1.0-6 haven_2.5.1
[70] cluster_2.1.4 fs_1.6.0 magrittr_2.0.3
[73] data.table_1.14.6 flextable_0.8.1 reprex_2.0.2
[76] googledrive_2.0.0 truncnorm_1.0-8 whisker_0.4.1
[79] hms_1.1.2 evaluate_0.20 xtable_1.8-4
[82] XML_3.99-0.10 jpeg_0.1-9 readxl_1.4.1
[85] gridExtra_2.3 compiler_4.2.0 mice_3.14.0
[88] writexl_1.4.0 crayon_1.5.2 htmltools_0.5.4
[91] later_1.3.0 tzdb_0.3.0 Formula_1.2-4
[94] geneplotter_1.74.0 lubridate_1.9.1 DBI_1.1.3
[97] dbplyr_2.3.0 Matrix_1.5-1 cli_3.4.1
[100] parallel_4.2.0 pkgconfig_2.0.3 getPass_0.2-2
[103] foreign_0.8-82 plotly_4.10.1 xml2_1.3.3
[106] foreach_1.5.2 svglite_2.1.1 annotate_1.74.0
[109] bslib_0.4.2 webshot_0.5.3 XVector_0.36.0
[112] rvest_1.0.3 callr_3.7.3 digest_0.6.31
[115] Biostrings_2.64.1 polylabelr_0.2.0 rmarkdown_2.20
[118] cellranger_1.1.0 htmlTable_2.4.1 gdtools_0.2.4
[121] lifecycle_1.0.3 jsonlite_1.8.4 viridisLite_0.4.1
[124] fansi_1.0.4 pillar_1.8.1 lattice_0.20-45
[127] KEGGREST_1.36.3 fastmap_1.1.0 httr_1.4.4
[130] survival_3.4-0 GO.db_3.15.0 glue_1.6.2
[133] zip_2.2.2 png_0.1-7 iterators_1.0.14
[136] bit_4.0.5 stringi_1.7.12 sass_0.4.5
[139] blob_1.2.3 latticeExtra_0.6-30 memoise_2.0.1